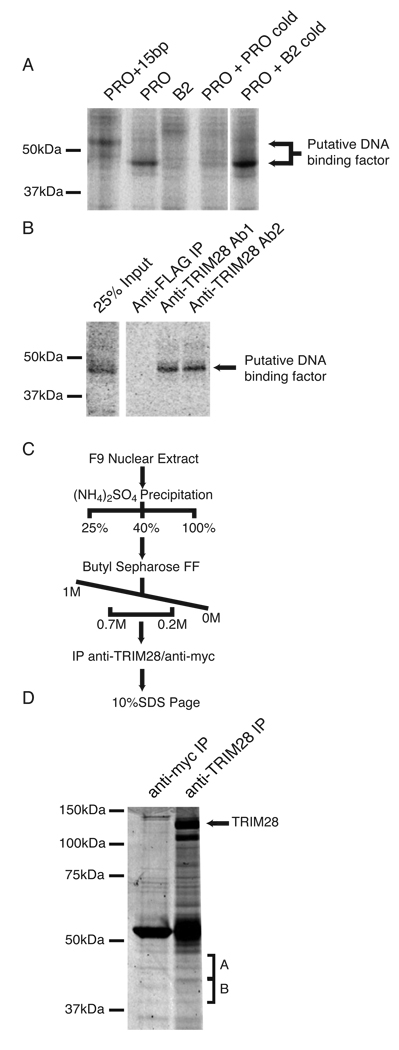
Figure 1. Identification of candidate PBSPro-DNA binding factors.
(A) F9 EC cell nuclear extract prepared and incubated with radiolabeled probes corresponding to PBSPro (PRO), PBSB2 (B2) or PBSPro with a 15 bp extension (PRO+15bp). Crosslinking was performed by exposure to UV light. Competition was achieved through the addition of 100 pmols of unlabeled probe corresponding to either PBSPro or PBSB2. (B) Crosslinked nuclear extracts were subjected to immunoprecipitation with anti-FLAG or two separate anti-TRIM28 antibodies8. (C) Purification scheme used to prepare samples for identification by mass spectrometry. (D) SDS PAGE of samples prepared by purification scheme outlined above, visualized by Coomassie stain. Bands excised for protein identification by mass spectrometry are labeled “A” and “B”.