Figure 2. AP2α suppresses C/EBPα promoter activity.
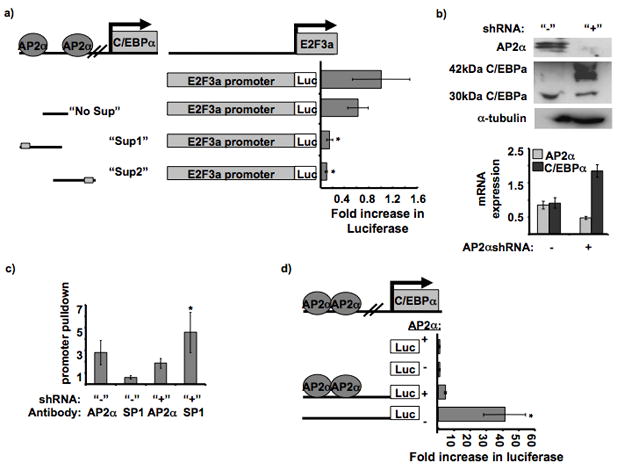
a. Effect of different parts of the C/EBPα suppressor sequence on E2F3a promoter activity. Diagram of the suppressor constructs and location of the AP2α binding sites. The different suppressor parts were cloned in front of the E2F3a promoter sequence in pGL3 basic. The luciferase values were normalized to the E2F3a promoter construct. The “Sup 1” and “Sup 2” constructs provided significantly less promoter activity than the “No Sup” and WT promoter constructs, * P = 0.0043 and 0.0048, respectively.
b. AP2α stable silencing and C/EBPα upregulation in SCC22B cell line. A short hairpin sequence targeting AP2α was cloned into pRS stable silencing vector. The western blot contains 300,000 untransfected SCC22B cells and 300,000 SCC22B cells transfected with the AP2α shRNA. The blot was probed with AP2α antibody,α-tubulin (internal control), and C/EBPα. Also, cDNA was prepared from RNA isolated from the respective cells. Quantitative RT-PCR data is shown for normalized AP2α and C/EBPα expression in SCC22B cells with and without AP2α shRNA. P = 0.0201 and 0.0121, respectively.
c. SP1 binding is inhibited by AP2α binding to the upstream C/EBPα sequence. Quantitative RT-PCR on C/EBPα promoter pulldown (−1423 to −1243 bp) with AP2α and SP1 antibodies in SCC22B cells before and after AP2α silencing. C/EBPαpromoter pulldown enrichment was normalized to the negative (no antibody) control. * P = 0.0451.
d. C/EBPα promoter activity before and after AP2α silencing. The promoter assay was performed in SCC22B cells (“+AP2α”) and SCC22B cells with downregulated AP2α (“−AP2α”). The promoter constructs contained −1423 bp of upstream C/EBPα sequence. Fold increase in luciferase activity was normalized to the negative control in each cell lysate (empty pGL3 basic). Promoter activity was significantly increased in the cell lysates with decreased AP2α expression. * P = 0.0237.
