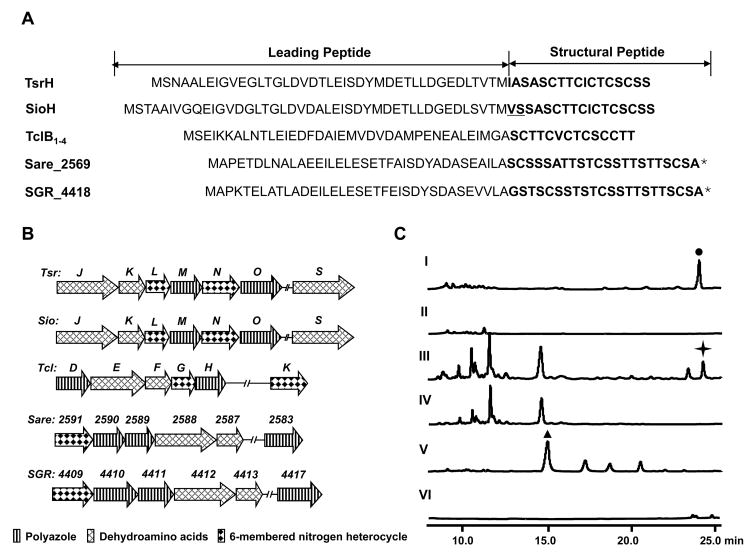
Figure 3.
Comparison and validation of the thiopeptide framework formation. (A) Peptide precursors for thiostrepton (TsrH), siomycin A (SioH), thiocillin I (TclB1–4), and two putative thiopeptides (Sare_2569 and SGR_4418). The SP sequences are labeled in bold (asterisk indicates the proposed SPs). The SP amino acids difference of SioH from TsrH is underlined. (B) Organization of the thiopeptide framework-forming genes identified from the producers of thiostrepton (tsr), siomycin A (sio), and thiocillin I (tcl), and two uncharacterized producers Salinispora arenicola CNS-205 (sare) and S. griseus subsp. Griseus NBRC 13350 (sgr). (C) HPLC analysis of the thiopeptide production in the wild type strain of the thiostrepton (solid dot) producer S. laurentii ATCC 31255 (I), tsrJ mutant S. laurentii strain SL1001 (II), wild type strain of the siomycin A (solid star) producer S. sioyaensis ATCC 13989 (III), sioO mutant S. sioyaensis strain SL2001 (IV), wild type strain of the thiocillin I (solid triangle) producer B. cereus ATCC 14579 (V), and tclE mutant B. cereus strain SL3001 (VI).