Abstract
The constitutive Cauliflower Mosaic Virus 35S promoter (CaMV 35S) is widely used as a tool to express recombinant proteins in plants, but with different success. We previously showed that the expression of an F-actin marker, GFP-talin, in Physcomitrella patens using the CaMV 35S promoter failed to homogenously label moss tissues. Here, we show a significant diminution of the GFP fluorescence in dark grown old moss cells and complete lack of labelling in newly differentiated cells. Furthermore, we demonstrate that stable moss lines harbouring a resistance cassette driven by the CaMV 35S are unable to grow in darkness in the presence of the antibiotic. In contrast to the CaMV 35S, the heat inducible promoter, hsp17.3B showed uniform expression pattern in all cells and tissues following a mild heat shock.
Key words: F-actin, constitutive/inducible promoter, heat shock, hsp17.3B, sodium salicylate
The CaMV 35S promoter is regularly used to drive the expression of recombinant proteins in plants. In angiosperms, several studies showed that reporter genes driven by this promoter display unequal tissue and developmental expression patterns.1–6 Several versions of the CaMV 35S promoter are being used in different plant species and an 800 bp HindIII-BamHI CaMV 35S fragment7 is the most commonly used in the moss Physcomitrella patens. CaMV 35S was intensively used to drive the expression of resistance genes, cDNAs as well as genomic sequences.8–10 Comparison between several true constitutive promoters in P. patens revealed that CaMV 35S has a much weaker activity as compared to the rice actin-1 and the maize ubiquitin-1 promoters.11,12 Although the expression of resistance genes using CaMV 35S is sufficient for the selection of transformed moss plants, the attempts to overproduce proteins of interests by CaMV 35S remain rather inefficient in moss. For instance, the complementation of abi3 mutant phenotype was not successful when 35S::ABI3 expression cassette was introduced in P. patens genome.13 Moreover, under standard conditions (16 h light/8 h dark), the expression of actin microfilament marker (GFP-talin) using CaMV 35S promoter, in 35S-GT line, resulted in detectable GFP fluorescence only in old cells while GFP levels were weak and undetectable in actively differentiating protonemal and meristematic cells.14 Our attempts to overcome this problem by using stronger promoters such as the rice actin-1,15 or the maize ubiquitin-1,16 promoters failed probably due to the high toxic amounts of produced GFP-talin (data not shown). These obstacles were overcomed by employing the inducible soybean hsp17.3B promoter12 to drive GFP-talin expression in HGT line.14
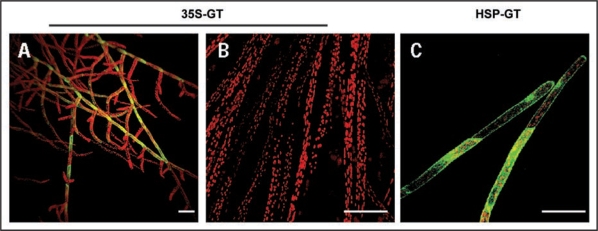
Caulonemal cells are the only protonemal cell type that can grow and divide in darkness in presence of a carbon source.17 To verify whether the expression of GFP-talin could be achieved in these newly differentiated cells, we monitored fluorescent labeling in dark grown caulonemal cells in 35S-GT and HGT lines. Whereas HGT cells remained unlabeled, a steady attenuation of GFP fluorescent signal was observed in the previously labeled tissue of 35S-GT line (Fig. 1A). Moreover, no GFP signal was detected in newly differentiated caulonemal cells (Fig. 1B). Whereas a short mild heat shock (1 h at 36°C) had no influence on labeling in 35S-GT line, a uniform GFP labeling was detected in all HGT newly developed cells (Fig. 1C).
Figure 1.
Confocal images showing GFP-talin expression in 35S-GT and HGT lines. (A–C) Merged confocal images displaying auto fluorescence of chloroplasts (in red) and fluorescence signal of GFP (in green). (A) 35S-GT grown under standard conditions. Ten days old, dark grown caulonemal cells of (B) 35S-GT and (C) HGT lines. The 35S-GT cells were imaged immediately after light exposure, whereas HGT cells were heat treated in darkness for 1 h at 36°C and imaged 16 hours after treatment. Bars: 100 µm.
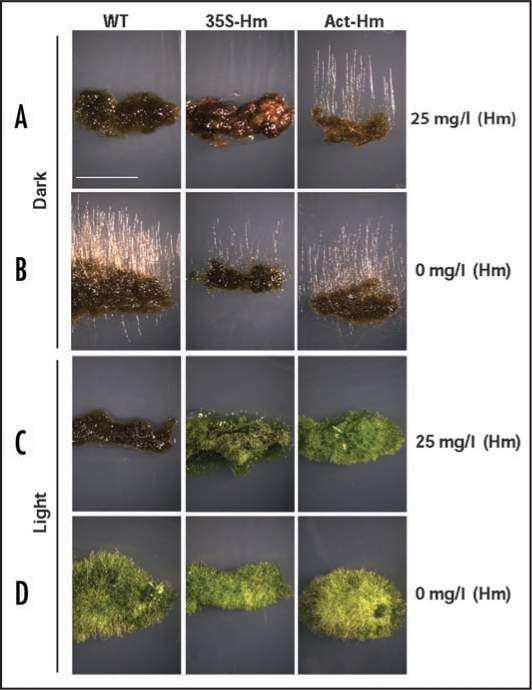
To further study the phenotypic consequences of the above-mentioned CaMV 35S-driven expression pattern, we compared the resistance to hygromicin of two moss lines both carrying the hygromicin phosphotransferase (hph) resistance gene driven either by CaMV 35S promoter (35S-HygroR) or the rice actin-1 promoter (Act-HygroR). We monitored caulonemal differentiation in 35S-HygroR and Act-HygroR lines grown under continuous darkness (Fig. 2A and B). In the presence of 25 mg/l hygromycin, differentiation and growth of 35S-HygroR caulonemal cells was drastically reduced compared to the one observed in Act-HygroR line. Under standard light conditions, colonies of both lines proliferated optimally and at similar rate regardless of the presence of hygromycin (Fig. 2C and D). Similarly, dark grown strains with integrated 35S-nptII (neomycin phosphotransferase II) expressing cassette could not produce etiolated caulonemal cells on medium containing G418 antibiotic (data not shown). These data indicate that resistance to antibiotic is seriously diminished during etiolating processes in lines harboring the resistance gene driven by CaMV 35S promoter.
Figure 2.
35S-Hm line is unable to grow in the dark when hygromycin is supplemented to the medium. Growth of WT (left), 35S-Hm (middle) and Act-Hm (right) lines using mediums supplemented with (A and C) or without (B and D) hygromycin (Hm, 25 mg/l). Six days old P. patens colonies were transferred to dark (A and B) or continued to grow in standard growing conditions (C and D) during ten days. Pictures were taken immediately after. Bars: 10 mm.
We conclude that moss etiolation make expression of transgenes from CaMV 35S promoter unattainable to the point of loss of transgene phenotypical characteristics. We may only speculate that the weak activity of CaMV 35S promoter is intrinsically more repressed in darkness or it is attributable to the lack of certain non-indentified transcription factor regulated by light. In moss cells, when expression of transgenes needs to be maintained in darkness, we therefore advocate the use of endogenous promoters (i.e., housekeeping genes) by gene conversion. Yet, if overexpression or ectopic expression is required the rice actin promoter is better choice than CaMV 35S promoter. Alternatively, when the permanent overexpression of proteins of interest causes adverse effect to P. patens, the use of the hsp17.3B inducible promoter is thus the best approach to date.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8541
References
- 1.Benfey PN, Ren L, Chua NH. Tissue-specific expression from camv 35S-enhancer sub- domains in early stages of plant development. EMBO J. 1990;9:1677–1684. doi: 10.1002/j.1460-2075.1990.tb08291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terada R, Shimamoto K. Expression of camv35s-gus gene in transgenic rice plants. Mol Gen Genet. 1990;220:389–392. [Google Scholar]
- 3.Yang NS, Christou P. Cell type specific expression of a camv 35s-GUS gene in transgenic soybean plants. Dev Genet. 1990;11:289–293. [Google Scholar]
- 4.Blumenthal A, Kuznetzova L, Edelbaum O, Raskin V, Levy M, Sela I. Measurement of green fluorescence protein in plants: Quantification, correlation to expression, rapid screening and differential gene expression. Plant Sci. 1999;142:93–99. [Google Scholar]
- 5.Sunilkumar G, Mohr L, Lopata-Finch E, Emani C, Rathore KS. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Molec Biol. 2002;50:463–474. doi: 10.1023/a:1019832123444. [DOI] [PubMed] [Google Scholar]
- 6.Halfhill MD, Millwood RJ, Rufty TW, Weissinger AK, Stewart CN. Spatial and temporal patterns of green fluorescent protein (GFP) fluorescence during leaf canopy development in transgenic oilseed rape, Brassica napus l. Plant Cell Rep. 2003;22:338–343. doi: 10.1007/s00299-003-0696-4. [DOI] [PubMed] [Google Scholar]
- 7.Guilley H. Transcription of cauliflower mosaic-virus DNA—detection of promoter sequences, and characterization of transcripts. Cell. 1982;30:763–773. doi: 10.1016/0092-8674(82)90281-1. [DOI] [PubMed] [Google Scholar]
- 8.Imaizumi T, Kadota A, Hasebe M, Wada M. Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell. 2002;14:373–386. doi: 10.1105/tpc.010388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezanilla M, Pan A, Quatrano RS. RNA interference in the moss Physcomitrella patens. Plant Physiol. 2003;133:470–474. doi: 10.1104/pp.103.024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finka A, Saidi Y, Goloubinoff P, Neuhaus JM, Zryd JP, Schaefer DG. The knock-out of ARP3a gene affects F-actin cytoskeleton organization altering cellular tip growth, morphology and development in moss Physcomitrella patens. Cell Motil Cytoskel. 2008;65:769–784. doi: 10.1002/cm.20298. [DOI] [PubMed] [Google Scholar]
- 11.Horstmann V, Huether CM, Jost W, Reski R, Decker EL. Quantitative promoter analysis in Physcomitrella patens: A set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol. 2004;7:4–13. doi: 10.1186/1472-6750-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saidi Y, Finka A, Chakhporanian M, Zryd JP, Schaefer DG, Goloubinoff P. Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat-shock promoter: A tool for plant research and biotechnology. Plant Mol Biol. 2005;59:697–711. doi: 10.1007/s11103-005-0889-z. [DOI] [PubMed] [Google Scholar]
- 13.Marella HH, Sakata Y, Quatrano RS. Characterization and functional analysis of abscisic acid insensitive3-like genes from Physcomitrella patens. Plant J. 2006;46:1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x. [DOI] [PubMed] [Google Scholar]
- 14.Finka A, Schaefer DG, Saidi Y, Goloubinoff P, Zryd JP. In vivo visualization of F-actin structures during the development of the moss Physcomitrella patens. New Phytol. 2007;174:63–76. doi: 10.1111/j.1469-8137.2007.01989.x. [DOI] [PubMed] [Google Scholar]
- 15.Mcelroy D, Blowers AD, Jenes B, Wu R. Construction of expression vectors based on the rice actin-1 (act1) 5′ region for use in monocot transformation. Mol Gen Genet. 1991;231:150–160. doi: 10.1007/BF00293832. [DOI] [PubMed] [Google Scholar]
- 16.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transg Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins GI, Courtice GRM, Cove DJ. Gravitropic responses of wild-type and mutant strains of the moss Physcomitrella patens. Plant Cell Environm. 1986;9:637–644. doi: 10.1111/j.1365-3040.1986.tb01621.x. [DOI] [PubMed] [Google Scholar]
- 18.Saidi Y, Domini M, Choy F, Zryd JP, Schwitzguebel JP, Goloubinoff P. Activation of the heat shock response in plants by chlorophenols: Transgenic Physcomitrella patens as a sensitive biosensor for organic pollutants. Plant Cell Environm. 2007;30:753–763. doi: 10.1111/j.1365-3040.2007.01664.x. [DOI] [PubMed] [Google Scholar]