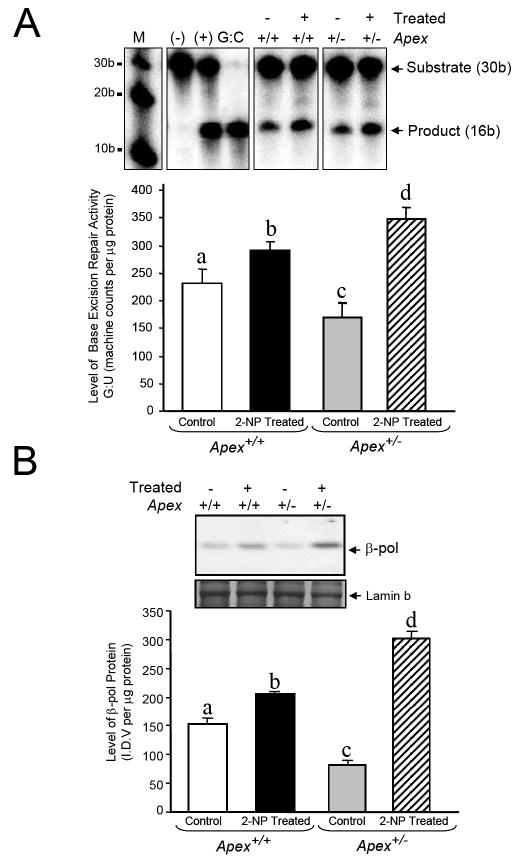
Fig. 5.
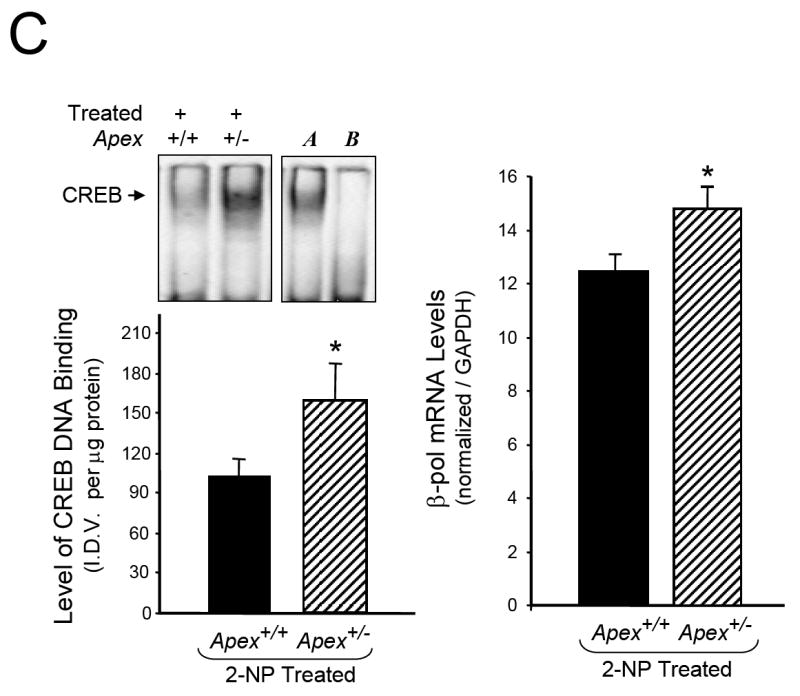
Effect of APE1/Ref-1 haploinsufficiency and 2-NP on G:U mismatch BER, DNA polymerase β expression and CREB DNA binding activity. (A) The in vitro G:U mismatch BER was conducted using nuclear extracts from liver of control and 2-NP treated Apex+/+ and Apex+/- mice. Base excision repair (BER) reaction products were resolved on a sequencing gel and visualized by autoradiography. Repair activity is visualized by the appearance of a 16b fragment. The relative level of BER was quantified using a Bio-Rad Molecular Imager® System. The data were normalized based on the amount of protein used in each reaction and expressed as machine counts per μg of protein. (B) The level of the 39-kDa β-pol protein in 200 μg of liver nuclear extract was determined by western blot analysis. (SDS-PAGE): the level of β-pol protein was normalized based on the amount of protein loaded on each gel. (C) The level of CREB DNA binding in 10 μg of liver nuclear extract was determined using EMSA and β-pol mRNA expression level as quantified using a real-time PCR and normalized with GAPDH Values represent an average (± S.E.M.) for data obtained from 5 mice in each group and are representative of separate identical experiments. (M): molecular weight standard; (-): negative control, G:U mismatch oligonucleotide incubated in the absence of nuclear extract and treated with HpaII restriction endonuclease; (+): positive control, G:U mismatch oligonucleotide incubated with recombinant β-pol and treated with HpaII restriction endonuclease; (G:C): positive control, G:C oligonucleotide incubated with nuclear extract and treated with HpaII restriction endonuclease; (I.D.V.): integrated density value corresponding to the level of β-pol protein and the level of CREB DNA binding as quantified by an AlphaInnotech ChemiImager™; Lane A & B: nuclear extracts incubated in the presence of 100X molar excess of mutated and unlabeled CRE sequence with β-pol flanking region respectively to confirm binding specificity; Lamin B protein level served as nuclear protein loading control. Values with different letter superscripts indicate significant differences at P < 0.05. (*): value significantly different from wildtype (Apex+/+) mice treated with 2-NP at P < 0.05. The picture depicts the representative sample from each group.