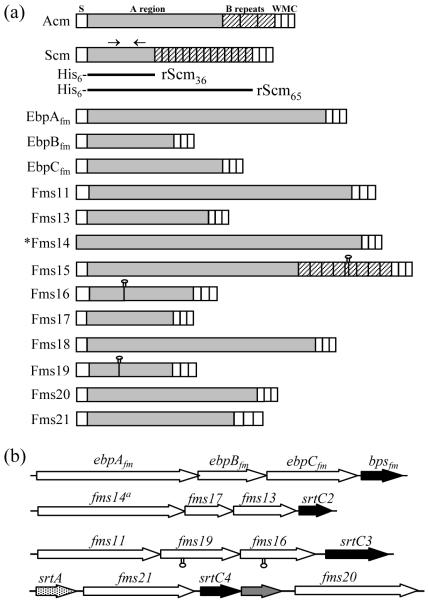
Fig. 1.
Domain organization of E. faecium CWA proteins with Ig-like folds. (a) S, signal peptide; W, cell-wall-spanning region; M, membrane anchor region; C, cytoplasmic tail with charged residues; Shaded A region, non-repetitive sequences; hatched B region, repetitive sequences; lollypop denotes presence of a stop codon due to a nucleotide substitution/deletion in Fms15, Fms16 and Fms19 of TX0016; solid black bars, His6-tag expressed regions of Scm (Fms10). Arrows above Scm show regions with predicted Ig-like folding. (b) Contiguously arranged ORFs encoding CWA proteins in E. faecium genome; srt, sortase. *Fms14 lacks an identifiable signal peptide, but has all the other characteristics of CWA MSCRAMMs.