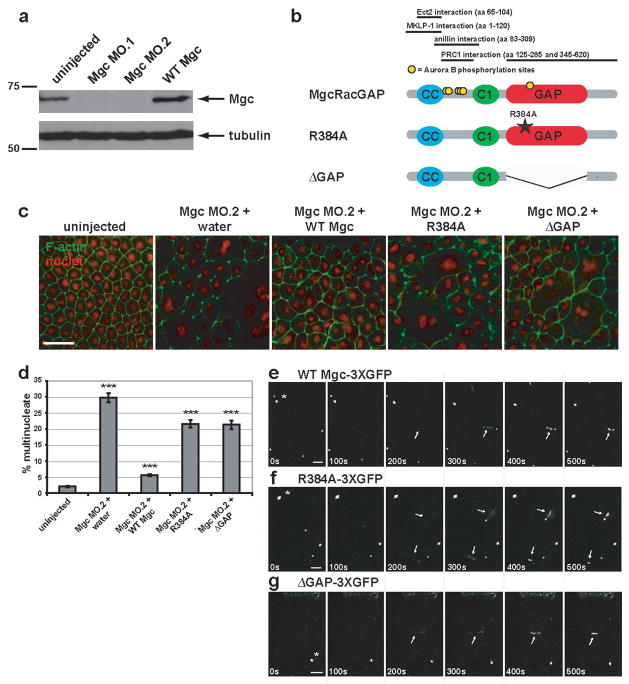
Figure 1. MgcRacGAP’s GAP activity is required for cytokinesis in Xenopus embryos.
a, Western blot showing morpholino knock-down of MgcRacGAP protein levels by Mgc MO.1 and MO.2 (see Fig. S1a for morpholino target sites) as compared to uninjected or WT Mgc mRNA-injected Xenopus embryo lysates harvested 24 h post-injection. The top panel was immunoblotted with antibodies to Xenopus MgcRacGAP, and the bottom panel with antibodies to α-tubulin as a loading control. b, Domain diagram of MgcRacGAP and its GAP-DEAD mutants, R384A and ΔGAP. CC = coiled coil domain, C1 = putative phorbol ester/diacylglycerol binding domain, GAP = GTPase activating domain. Regions that interact with Ect2, MKLP-1, anillin, or PRC1 as well as sites phosphorylated by AuroraB are noted. c, Embryos were injected with MO.2 along with either water, WT Mgc mRNA, Mgc R384A mRNA, or Mgc ΔGAP mRNA. Embryos were fixed at 24 h post-injection and stained with propidium iodide to reveal the nuclei (red) and Alexa 488-phalloidin to visualize F-actin (green). Scale bar, 50 μm. d, The percent of multinucleate cells was quantified for gene replacement embryos from three independent experiments. Mean ± SE. Uninjected, n = 10 embryos; Mgc MO.2 + water, n = 10; Mgc MO.2 + WT Mgc, n = 11; Mgc MO.2 + R384A, n = 11, Mgc MO.2 + ΔGAP, n = 11. ***p < 0.005. e–g, Frames from time-lapse movies showing the localization patterns of WT Mgc-3XGFP (e), R384A-3XGFP (f), or ΔGAP-3XGFP (g). Asterisks indicate midbodies (from previous cell divisions), which are extremely bright. Arrows indicate the localization of the constructs at the closing cytokinetic apparati. The mutants are detectable at the equator at least as soon as WT MgcRacGAP (see 200s time point). Scale bars, 20 μm.