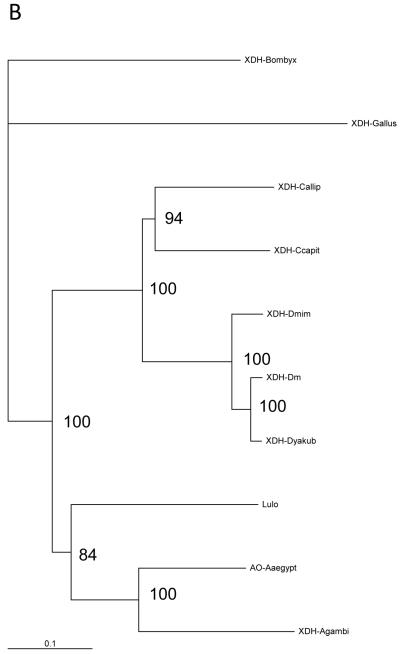
Fig. 1.
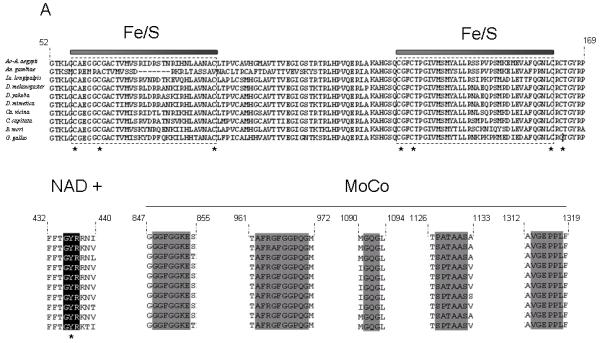
(a) Partial structure-based alignment of the amino acid sequence of Lulo-XDH (Accession no AM887864) and some other insect xanthine dehydrogenases/aldehyde oxidade: Anopheles gambiae XDH (Accession no AAO14865); Aedes aegypti aldehyde oxidase (Accession no EAT46105); Drosophila melanogaster XDH (Accession no CAA68409); Drosophila mimetica XDH (Accession no AAQ17532); Drosophila yakuba XDH (Accession no AAQ17528); Calliphora vicina XDH (Accession no CAA30281); Ceratitis capitata XDH (Accession no AAG47345); Bombyx mori XDH1 (Accession no BAA21640). Conserved functional areas of the enzyme are 2Fe/2S iron-sulfur domains (boxed), NAD+-binding site (black shade) and the carboxy terminal MoCo molybdenum cofactor (grey shade). (b) Phylogenetic tree of XDH sequences among insect species and Gallus gallus (domestic chicken) XDH obtained by the Neighbour Joining method. The scale bar represents 0.1% divergence. The number in the tree branches indicates the bootstrap value from 1000 interactions.