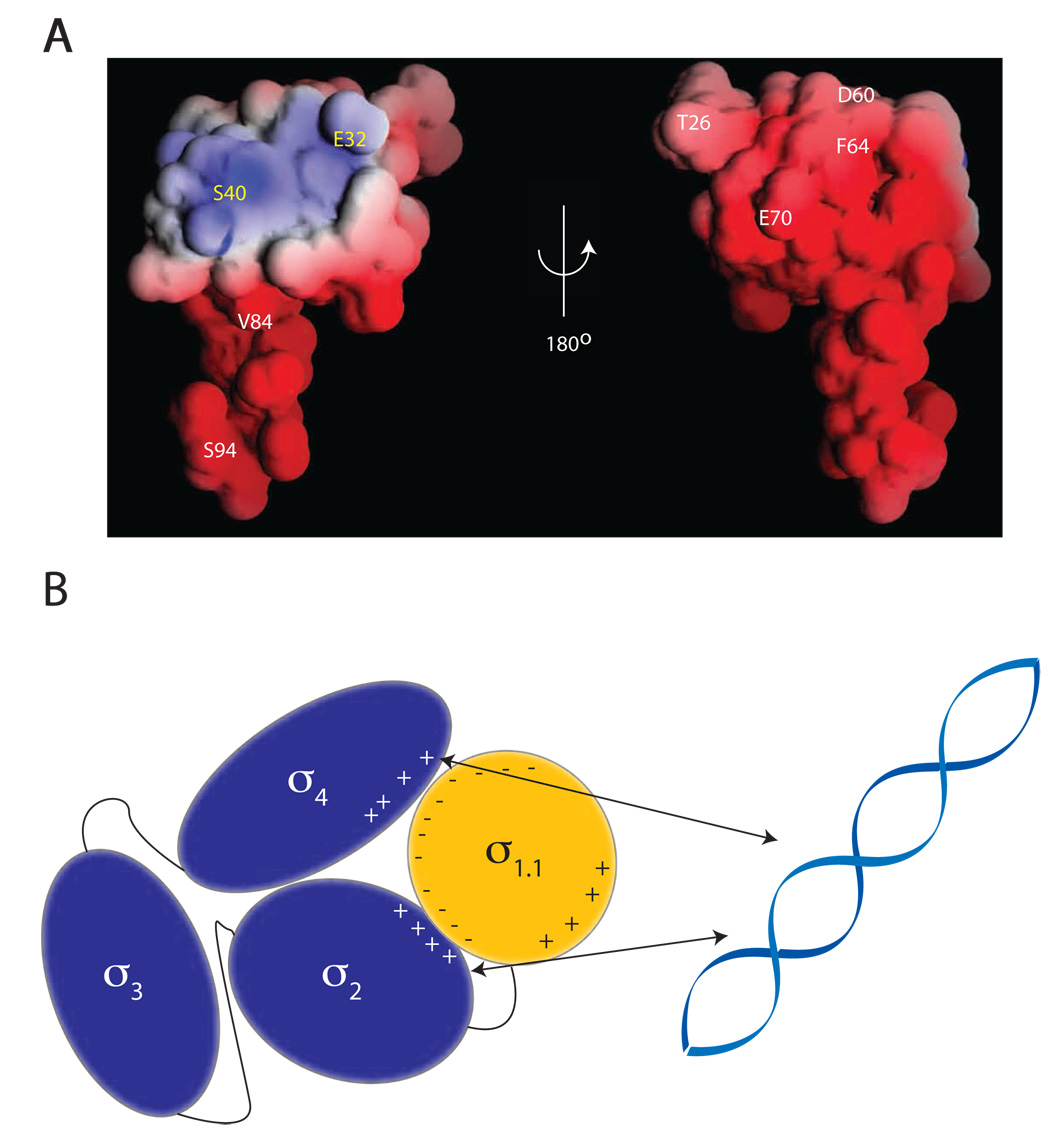
Figure 6. Proposed model of DNA binding inhibition by σ1.1.
(A) Crosslinker attachment sites are indicated on the electrostatic surface map of σ1.1 generated using the GRASP program. Indicated in white are the sites that were shown to make interdomain crosslinks. Indicated in yellow are the sites from which no interdomain crosslinking was observed. (B) Schematic showing the compaction model of σA autoinhibition. The negative surface of σ1.1 is capable of forming crosslinks to the DNA binding domains σ2 and σ4. It is thus likely that σ1.1 organizes the σ factor into a compacted structure that is incapable of binding DNA. σ2 forms crosslinks to σ4, indicating that these two domains must also be in close proximity.