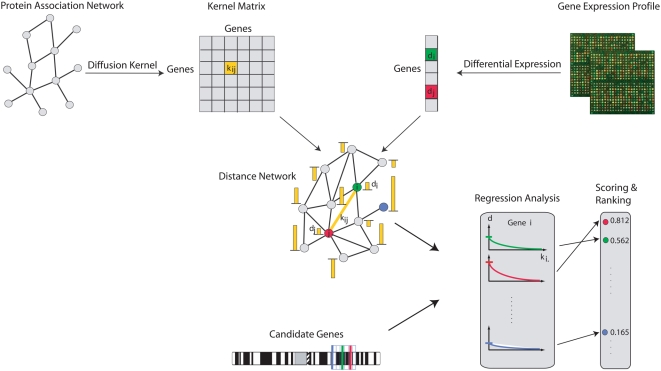
Figure 1. Overview of the method.
(1) From a protein association network a global distance network is computed based on a kernel method (e.g., the Laplacian exponential diffusion kernel). (2) A disease related microarray experiment is required from which the differential expression levels of all genes in the network are determined. (3) The differential expression levels of the genes are mapped on the global distance network. (4) A set of candidate genes is required (e.g., from a linkage study). (5) For each candidate gene its differentially expressed neighborhood is identified by a regression analysis. (6) Based on the regression analysis each candidate gene receives a score. (7) The candidate genes are ranked by their scores.