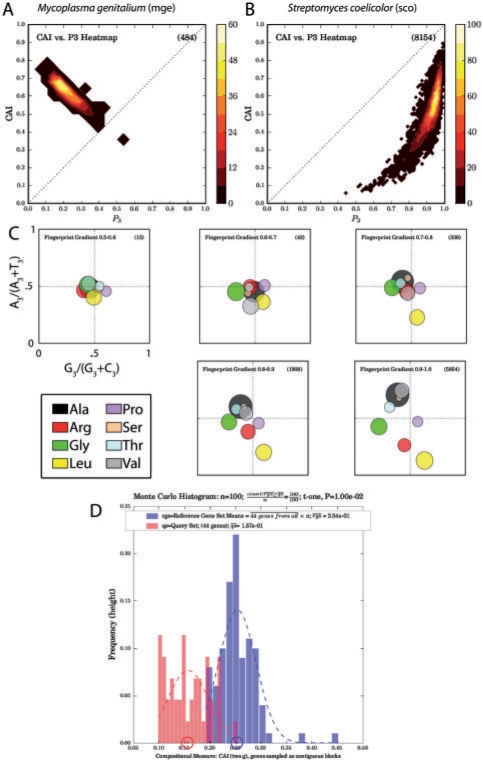
Fig. 1.
Examples of CodonExplorer output. (a and b) Plots of CAI (a measure correlated with expression) against third position GC content for two genomes with extreme codon bias, M.genitalium and S.coelicolor. (c) Fingerprint plots broken down by ranges of per-gene GC content: codon usage is more biased at higher GC content. (d) Monte Carlo histogram of CAI values for the Salmonella pathogenicity island 2 (SPI-2), which differs significantly (P=0.01) in average CAI values from the genome as a whole using a t-test to compare the actual mean to a distribution of means of other genes: these should be approximately normally distributed due to the Central Limit Theorem.