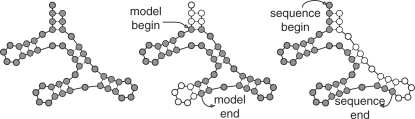
Fig. 1.
Comparison of local alignment types. Left: global alignment; filled circles indicate observed residues in an RNA structure, which can be thought of as a binary tree. Center: subtree method of local RNA structural alignment. Whole domains of the RNA structure may be skipped (open circles indicate consensus positions without aligned sequence residues), but the observed alignment satisfies all expected structural constraints: if a residue is aligned to a pair state, another residue will be aligned to form a base pair. Right: truncated sequence method of local RNA structural alignment, where the observed sequence may begin and end anywhere with respect to the consensus RNA structure. Aligned residues may be base-paired to positions that are missing from the alignment.