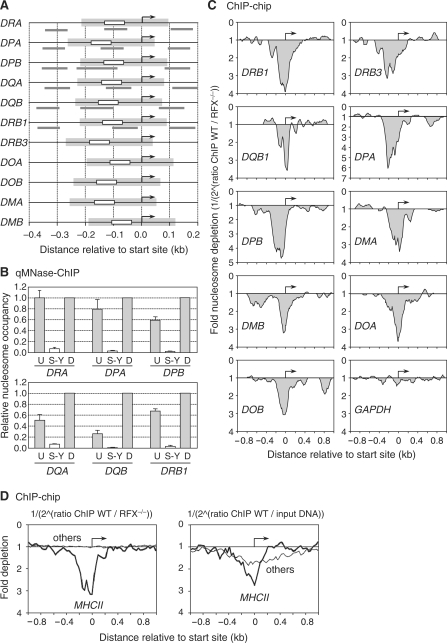
Figure 4.
Nucleosome eviction from the S-Y/TSS region occurs at all MHCII genes. (A) MHCII promoters were aligned relative to their TSSs (arrows). Positions of the S-Y modules (white boxes), predicted NFRs (gray boxes) and amplicons (gray lines) used in panel B are indicated. The sizes and positions of the predicted NFRs were extrapolated from the NFR that was mapped at the DRA promoter. (B) Nucleosome occupancy in the vicinity of the S-Y/TSS regions of the HLA-DRA, HLA-DPA, HLA-DPB, HLA-DQA, HLA-DQB and HLA-DRB1 genes was measured by qMNAse-ChIP in Raji B cells. Primer pairs were situated upstream of (U), within (S-Y) and downstream of (D) the S-Y/TSS NFRs. Results were normalized with respect to MNAse treated genomic DNA, expressed relative to the value obtained at the downstream position, and show the means and standard deviations derived from three independent experiments. Amplicons used are depicted in panel A and indicated in Supplementary Table 1. (C) Nucleosome occupancy at the indicated MHCII genes was analyzed by ChIP–chip using antibodies directed against unmodified histone H3 and MNAse treated chromatin from wild-type Raji B cells (WT) and RFX-deficient B cells (RFX−/−). The results are represented as the fold nucleosome depletion in WT relative to RFX−/− cells. The NFRs and TSSs are indicated by gray shading and arrows, respectively. (D) The average nucleosome density observed at the TSS of MHCII genes (thick lines) was compared with that found at 50 other genes present on the array and expressed most strongly in B cells (thin profiles). Results represent the average fold depletion observed in ChIP samples from wild-type Raji cells (WT) relative to ChIP samples from RFX-deficient (RFX−/−) cells (left) or input DNA (right). The TSSs are indicated by arrows.