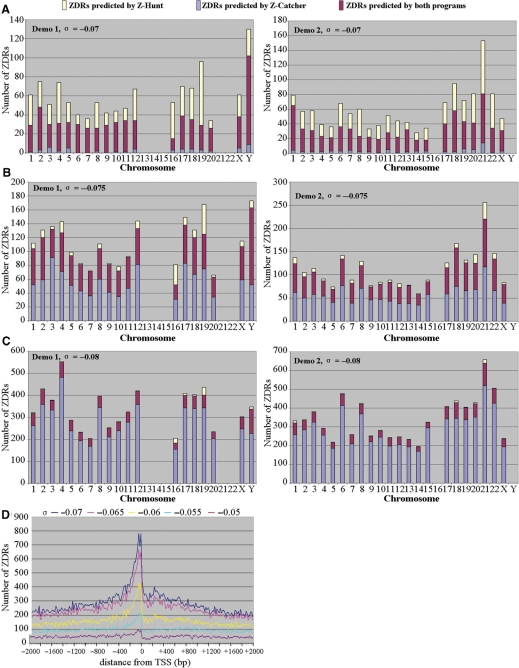
Figure 1.
Predictions of ZDRs by Z-catcher. (A–C) Comparison of ZDRs identified by Z-catcher and Z-hunt. See text for details. Note that the randomly selected sequences from chromosomes 13, 14, 15, 21 and 22 in Demo 1 contained undefined bases, which prevented in silico analyses. The same applied to sequences from chromosomes 16 and Y in Demo 2. (D) Distribution of ZDRs at various σ levels around TSS in the human genome. All 103 542 representative TSSs of transcription units in the human genome from the Database of Transcriptional Start sites (DBTSS release 5.2.0) were selected. Flanking regions of TSSs 2000 bp upstream and 2000 bp downstream were extracted and subjected to Z-catcher, with σ values ranging from –0.070 to –0.050. The distances between the predicted ZDRs and their corresponding TSSs were calculated and plotted.