Abstract
The discovery of microRNAs (miRNAs), almost 10 years ago, changed dramatically our perspective on eukaryotic gene expression regulation. However, the broad and important functions of these regulators are only now becoming apparent. The expansion of our catalogue of miRNA genes and the identification of the genes they regulate owe much to the development of sophisticated computational tools that have helped either to focus or interpret experimental assays. In this article, we review the methods for miRNA gene finding and target identification that have been proposed in the last few years. We identify some problems that current approaches have not yet been able to overcome and we offer some perspectives on the next generation of computational methods.
INTRODUCTION
A novel post-transcriptional silencing process was discovered at the turn of the century. It is elicited by tiny endogenous RNAs called microRNAs (miRNAs). miRNAs are a large class of small non-coding RNA molecules that have early on been recognized to be numerous and phylogenetically extensive (1,2). Many of these molecules originate from non-coding genes which produce mature transcripts of ∼22 nt in length and are thought to function primarily as antisense regulators of other RNAs (3). A detailed history of the discovery of these regulatory molecules is available in (4).
The initial members of the miRNA class of non-coding RNAs were lin-4 and let-7 of Caenorhabditis elegans. They were termed heterochronic or small temporal RNAs (1,2) because all known instances seemed to be involved in controlling the timing of larval development. Most known miRNAs are very well conserved in close species and some can be found across very large taxonomic groups, notably let-7 of C. elegans (5).
The aim of this article is to offer an overview of the open problems, current methods and future perspectives in the field of miRNA computational biology, with an emphasis on the shortcomings of the several approaches that have been used so far as well as a description of the latest insights that may inspire the development of the next generation of computational methods for miRNA gene finding and target prediction.
BIOGENESIS AND FUNCTION
miRNA genes are frequently expressed individually, but many exist in clusters of 2–7 genes with small intervening sequences. Experimental results suggest that they are expressed co-transcriptionally, which indicates that they are under control of common regulatory sequences (2,6,7).
Other miRNA genes are excised from the introns of protein-coding genes (8,9) introns and exons of non-coding genes (10), or even from the 3′-UTR of protein-coding genes (11). In mammalian genomes, it is also possible to find miRNAs in repetitive regions, and some studies suggest that transposable elements may be involved in the creation of new miRNAs (12).
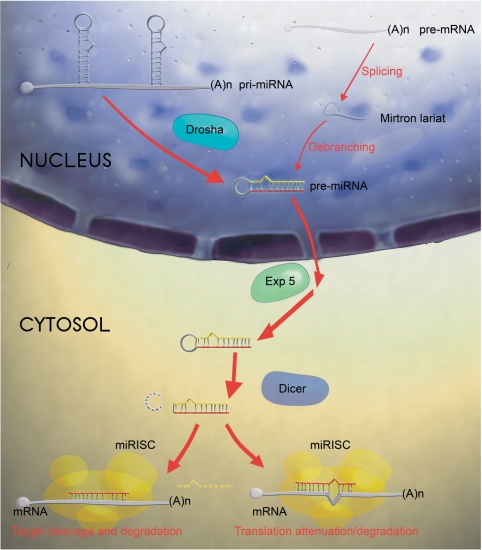
miRNA biogenesis in animals is a two-step process (6), as shown in Figure 1. The nascent transcript, which is several hundred nucleotides long, is called primary miRNA (pri-miRNA). Although some miRNAs are transcribed by RNA pol III (13), most rely on RNA pol II (4,11), therefore pri-miRNAs can be subjected to elaborate transcriptional control.
Figure 1.
The miRNA biogenesis in metazoans. The figure shows two major pathways for metazoan miRNA biogenesis. The pri-miRNA is indicated as a polycistronic transcript. The stem–loops are cleaved by Drosha in the nucleus giving rise to the pre-miRNA. Alternatively, the pre-miRNA can originate from a particular kind of intron—the mirtron. The pre-miRNA is shown with a red strand (the mature miRNA) and a yellow strand (the miRNA*). The pre-miRNA is then exported by Exp5 and processed by Dicer in the cytosol. The red strand of the resulting duplex is integrated in the miRISC and the yellow strand is degraded. Depending on the degree of complementarity to the target site, the silencing complex will either cleave the mRNA inducing immediate degradation or promote translational attenuation. The mechanism of translational attenuation can also subsequently promote target degradation.
In a first step, the primary transcript is processed in the nucleus by a multiprotein complex (Microprocessor) containing an enzyme called Drosha (15) to give rise to the ∼70-nt long miRNA stem–loop precursor (pre-miRNA) which is then exported to the cytoplasm. Secondary structure, rather than primary sequence, seems to be a critical feature for Drosha substrate recognition (16), however it is not known how this enzyme discriminates pre-miRNAs from the great variety of cellular RNA stem–loops. It is known, however, that efficient processing by Drosha is dependent upon the presence of unstructured regions flanking the stem–loop (17). The nuclear export is elicited by a complex of Exportin 5 (Exp5) and Ran-GTP which selectively binds pre-miRNAs while also protecting them from exonucleolytic digestion (18,19).
In the cytoplasm, a second step takes place where the pre-miRNA matures into a ∼21-nt long miRNA:miRNA* duplex, with each strand originating from opposite arms of the stem–loop. The cleavage is produced by the action of an enzyme called Dicer (20), which recognizes the double-stranded stem (21).
In general, the miRNA strand is then integrated in a ribonucleoprotein complex known as the miRNA-induced silencing complex (miRISC) or miRNA-containing ribonucleoprotein particles (miRNPs) and the miRNA* is degraded (2). Sometimes both strands can be detected (22), in which case the miRNA* designates the less predominant form of the mature miRNA.
Studies have shown that the intermediate miRNA duplexes exhibit a biased internal strand stability not only due to base pair composition but also to structural features like mismatches or bulges (23). These destabilizing elements are thought to facilitate unwinding of the duplex and subsequent integration in the silencing complex. The strand that is less stable on its 5′ end is preferably loaded onto miRISC (24). When both ends exhibit similar stability, each strand is selected for integration with similar frequency (22). Other studies, however, have suggested the existence of additional strand selection determinants (25,26).
miRNA biogenesis in plants follows a similar process, but the miRNAs seem to be fully matured into a single-stranded miRNA before being exported to the cytoplasm by an homologue of Exp5 termed HASTY (HST) and integrated onto the silencing complex, which partially explains why intermediate forms of plant miRNAs are only rarely detected (27,28).
All maturation steps of plant miRNAs are processed by Dicer-like proteins. The predicted miRNA precursors in plants are much more variable in size than those of animals, ranging from around 60 to a few hundred nucleotides, whereas those in animals are typically ∼70-nt long (29).
Given the stepwise process by which miRNAs are matured, and hence the diverse opportunities of regulation at each step, one can expect to find regulatory mechanisms at this level and, in fact, there is some evidence of post-transcriptional control of miRNA expression (16,30,31).
As mentioned, some miRNAs originate from the introns of other genes, usually being located in the same strand, which suggests that they are transcribed with the host genes and subsequently excised (8–10,32). Studies show that the expression of these miRNAs and their host genes is coupled, indicating a possible mechanism by which a protein and a miRNA are coordinately expressed (7,28), presumably as part of a common biological process. However, some intronic miRNAs occur in antisense orientation and may thus be transcribed under the influence of an independent promoter (16,33).
An alternative pathway for intron-derived miRNAs has been recently identified in animals (34). These introns, termed mirtrons, bypass Drosha processing and exhibit structural features similar to those of pre-miRNAs, thus entering the miRNA biogenesis pathway at the end of the first step. Unlike other intron-derived miRNAs which are excised from unspliced transcripts (35), mirtrons are dependent on the splicing machinery for maturation.
miRNAs in animals are thought to act primarily as translational repressors by pairing with specific partially complementary 3′-UTR regulatory elements on mRNAs (36), although target sites in the coding region and 5′-UTR can also be functional (37,38). Another major miRNA silencing mechanism in animals leads to target mRNA destabilization through a cleavage-independent process with a clear impact on transcript level (39,40). Some authors have suggested that miRNAs may have either a negative or positive regulatory effect (3). In fact, recent evidence indicates that positive transcriptional regulation can be produced by miRNAs that target sites in promoter regions by an otherwise unknown mechanism (41). Moreover, there are reports that in some circumstances and in certain cell-types, miRNAs can also enhance translation (42).
Plant miRNAs, on the other hand, frequently cleave and thus induce immediate degradation of the target mRNAs and are often almost perfectly complementary to sites in the coding region (43), as well as in the 3′-UTR (44), and even in the 5′-UTR (45). However, some of these target sites may only be present after mRNA maturation since they span intron/exon boundaries (46). It is also important to note that, a priori, nothing seems to prevent miRNAs from regulating RNAs other than mRNAs. They may also bind and regulate non-coding RNAs, perhaps even other miRNAs (3). This possibility is illustrated by a study done with Arabidopsis thaliana, which suggests that miRNAs may bind fake targets in other non-coding RNAs thereby establishing a mechanism of negative regulation of miRNA activity (47).
Unsurprisingly, some large DNA viruses have evolved ways to explore the RNA silencing machinery of the host by coding for miRNAs (48). These viral miRNAs can be expressed either individually or in clusters from pol II or pol III promoters. Interestingly, these miRNAs show no resemblance to other viral miRNAs, nor to the miRNAs of the host.
There are many computational problems associated with the miRNA world. The most important and also those that have drawn more attention are miRNA gene finding and target prediction. We discuss these problems in the two following sections and we present a list of online resources in the Supplementary data.
miRNA GENE FINDING
Large-scale experimental approaches to miRNA gene finding met with some difficulties in the beginning, illustrated by the fact that these regulators escaped detection for so long. The short length of miRNAs and their ability to act redundantly, or to have only a subtle phenotypical impact imposes a limitation to the use of mutagenesis and other conventional genetics techniques (3). Direct cloning, on the other hand, may not detect miRNAs that have very low expression levels or that are expressed only in specific conditions and cell types. This is partially mitigated by the use of deep-sequencing techniques which nevertheless require extensive computational analyses to distinguish miRNAs from other non-coding RNAs of similar size (49). It is clear, therefore, that computational approaches are essential for a more thorough catalogue of miRNA genes in sequenced genomes (8,50).
Conventional in silico gene finding approaches are of limited use since miRNAs and non-coding genes in general do not exhibit the characteristic statistical properties of coding regions due to codon usage. The same can be said for homology-based searches in the absence of a clear evolutionary model for these genes. Obtaining such a model is particularly difficult due to the comparatively small size of the precursor and mature sequences.
The different characteristics of miRNAs in animals and plants have justified different approaches and, therefore, we discuss the methods developed for the two cases separately.
Computational approaches to miRNA gene finding in animals
Lee and Ambros (1) established the paradigm of what would become the typical strategy for miRNA gene search. According to these authors, future miRNA genes ought to share some features with lin-4 and let-7 of C. elegans, namely the expression of a mature RNA sequence of the appropriate length (∼22 nt), which should have its origin in intergenic sequences and be processed from a stem–loop precursor transcript of around 65 nt in length. Furthermore, there should be extensive sequence similarity with orthologues in closely related species.
These observations prompted the adoption of several criteria for the annotation of novel miRNA genes (51). First, expression criteria establish that new miRNA genes should be supported by experimental evidence that detects the ∼22-nt RNA transcript, or that these small molecules should be found in cDNA libraries. Second, at least one of the following biogenesis criteria should be met: (i) the mature miRNA should be included in one arm of a predicted minimum free energy fold-back precursor structure with extensive base-pairing in the miRNA region which should not contain any large internal loops or bulges, especially asymmetric bulges; (ii) the fold-back structure should be phylogenetically conserved; and (iii) the precursor should be shown to accumulate in organisms with impaired Dicer function. Expression criteria need not be met in the case of obvious homologues.
It is clear that expression criteria alone are not sufficient for a confident annotation since they cannot distinguish miRNAs from other cellular RNAs with approximately the same size, or from spurious degradation products of other RNAs. On the other hand, the fact that expression evidence cannot be found does not necessarily exclude a candidate due to the limitations of experimental methods.
Known miRNA precursors have a typical stem–loop secondary structure which is essentially conserved amongst metazoa but heterogeneous within plants. Some of the first miRNA gene searches were carried out considering both this typical secondary structure and structure/sequence conservation between two closely related species (C. elegans and C. briggsae) (1). However, it soon became clear that there were much more conserved stem–loops than miRNA genes, and additional criteria had to be put in place if we were to identify good candidates (4).
Moreover, although a significant fraction of known miRNAs seems to be very well-conserved phylogenetically, this may reflect the bias of the search procedures used so far, which privilege phylogenetic conservation in order to validate miRNA candidates. It may also illustrate a general limitation of current computational approaches which can only predict candidates which resemble previously identified miRNAs (52). Furthermore, strong conservation may be a sign of the existence of multiple conserved target sites which would constitute an overwhelming selective force against mutation. As more organisms are being analysed, more miRNA genes are identified and an increasing number is shown to be lineage or species specific (53).
Despite the caveats, the annotation criteria have inspired most current computational methods for miRNA gene finding. Therefore, many tools share the same overall strategy but use different approaches to phylogenetic conservation, and different features to identify good stem–loop candidates. These methods can thus be distinguished roughly by the way they identify the initial candidate set, the structural criteria they use to further restrict precursor candidates, the conservation criteria they adopt and any additional filters they may implement. We refer to these approaches as filter-based methods. Later approaches use conventional machine learning methods that try to generalize from a positive set of previously known miRNAs and a negative set of stem–loops presumed not to be miRNA precursors. Target-centered approaches use a putative set of miRNA targets derived from conservation analyses which are then used to seek new miRNAs. Mixed approaches use a combination of computational tools and high-throughput experimental procedures. Finally, homology-based searches try to identify stem–loops similar to previously identified pre-miRNAs that may have been missed by ab initio methods.
Filter-based approaches
Early miRNA gene finding methods, summarized in Table 1, focused on the identification of small high-quality sets of conserved miRNA candidates which would have a better chance of being experimentally confirmed as true miRNAs. One of these methods, described in (50), identified several new miRNAs in C. elegans. An initial candidate set of imperfect stem–loops obtained from all repeats-masked intergenic regions of the genome of C. elegans was filtered according to criteria that accounted for matches, mismatches and gaps on the stem region, as well as GC content, MFE (minimum free energy) and the occurrence of multi-loops. The cut-offs for these parameters were chosen to reflect the characteristics of previously known miRNAs from the studied organism. Most of the filtering was achieved with the conservation criterion that required homologous stem–loops on two additional genomes. It is interesting to note that, from a universe of 61 known genes, only 29 out of 39 C. elegans miRNAs included in the initial set passed the structural criteria and not more than six miRNAs were conserved in two additional genomes, illustrating the emphasis on specificity rather than sensitivity.
Table 1.
Comparison of some filter-based approaches to miRNA gene finding in animals
| Initial set | Structural criteria | Conservation criteria | Additional filters | |
|---|---|---|---|---|
| Grad et al. (50) | Stem–loop structures in repeats-masked intergenic regions | MFE, GC content, matches, mismatches, gaps and occurrence of multi-loops | Homologous stem–loops transitively identified in two additional genomes | Hairpins containing short repeats or with low quality structure are eliminated |
| MirScan (8) | Folded structures identified sliding a 110-nt window along the genome | Number of bp, MFE, no overlap with repeats, no skewed base composition | Homologous stem–loops identified in an additional genome | Log-odds score for several features of the miRNA region of the stem–loop |
| Berezikov et al. (54) | Regions exhibiting a typical conservation pattern identified using phylogenetic shadowing | Only highly probable stable stem–loops are retained | Implicitly considered in the initial set | |
| MiRSeeker (9) | Aligned non-coding non-annotated regions from two species | Metrics involving length of longest stem–arm, MFE, internal loops, asymmetric loops and bulges applied to predicted structures in aligned regions | Typical divergence pattern |
Another approach, also published in 2003, makes some improvements on sensitivity. This method called MiRscan (8), produced an initial set of candidates by scanning the genome of C. elegans with a sliding-window of 110 nt. The regions were folded and filtered according to more permissive structural criteria. Potential homologues were sought in C. briggsae sequences and only conserved hairpins were retained, yielding a total of ∼36 000 candidates. With this procedure, 50 of the 53 miRNAs known at the time to be conserved in both species were recovered. Using these 50 miRNAs and the background set of over 36 000 hairpins, the authors developed a sophisticated log-odds scoring scheme that considered several features of the mature miRNA portion of the stem-loop. All candidate hairpins were scored and ranked according to this scheme. However, MiRscan was still not able to recover more than half of the previously known C. elegans miRNAs from the top scoring candidates.
The authors would later improve this method with MiRscanII (55) which, in addition to the features considered by MiRscan, took into account the presence of conserved motifs and blocks of sequence conservation up- and downstream of the predicted stem–loop precursors, presumably involved in transcriptional regulation. The authors observed that independently transcribed miRNA genes in C. elegans contained a well-conserved motif upstream of the stem–loop, with respect to homologous sequences in C. briggsae, and used it as an additional feature. Similar upstream motifs were found in Homo sapiens, Mus musculus, and Drosophila melanogaster.
An approach described in (54) also considers conservation around the precursor region and was used in the search for mammalian miRNAs. In this study, the authors could not identify clearly conserved motifs in the flanking regions immediately adjacent to the pre-miRNA stem–loops, but they were able to observe a distinctive pattern of diminishing conservation that was used as a characteristic profile aiding the search for miRNA genes.
The method miRseeker (9) represents the first attempt to identify conserved stem–loops due to selection, and not as an artefact of considering genomes that are not sufficiently distant. The authors aligned the non-annotated intergenic and intronic sequences of the genomes of D. melanogaster and D. pseudoobscura. The conserved regions were then folded in order to identify and score potential stem–loop structures. The evaluation of the hairpins considered the length of the longest stem arm and its MFE, as well as a set of metrics penalizing internal loops, particularly asymmetric loops and bulges. By analysing a reference set of known miRNA genes of two drosophilid species, the authors derived a typical divergence pattern. In general, divergence was observed in the terminal loop, or in either one of the stem arms. A good miRNA candidate should exhibit a pattern such that divergence occurs in at most one stem arm, and the mutation rate at the stem arm should not exceed that seen in the terminal loop. This is justified by the fact that mutations in the terminal loop have a lesser impact on pre-miRNA structure and identity and, consequently, its processing efficiency and target specificity, than mutations on the stem arm.
The methods described so far and variations thereof have been able to recover a substantial part of the known miRNAs and have been useful in identifying several new regulators (30,56,57). Some of these methods have benefited from a growing number of sequenced species allowing more extensive and sophisticated studies of conservation patterns (58,59). However, they have failed to produce a set of rules capable of recovering all known miRNAs without leading to too many false positives. Additionally, they are critically dependent on conservation criteria to attain reasonable levels of specificity. This approach effectively prevents the identification of non-conserved candidates, and makes several assumptions in the absence of a clear evolutionary model for these structures.
Machine learning methods
One attempt to use a single-genome approach to miRNA gene finding was ProMIR (60). The initial set of candidates are stem–loops that are present on human ESTs, therefore restricting the search to sequences with verified expression. Candidate stem–loops were filtered using very permissive structural criteria concerning stem length, loop size and MFE. This probabilistic method relies on an HMM (hidden Markov Model) that models characteristics of the stem portion of the stem–loop viewed as a paired sequence. These characteristics concern the pattern of base-pairing and the location of the mature miRNA. The positive training set consisted in all known human pre-miRNAs and the negative set corresponded to 1000 extended stem–loops randomly extracted from the human genome. A stem–loop is found to be a good pre-miRNA candidate if it contains a sequence with probability of being a mature miRNA above a certain threshold. However, the candidate set was still too large and additional filters had to be used, including the assessment of the statistical significance of the predicted secondary structure, and the verification of a decaying conservation pattern in the regions flanking the putative pre-miRNA by comparison to other vertebrate genomes, as done in (54).
The first successful single-genome approach came from a method developed to identify miRNAs in viral genomes (48). Using conservation criteria in this case is not an option as most viral pre-miRNAs show no detectable conservation with respect to either other viral pre-miRNAs or to the precursors of the infected host. The method starts by identifying robust stem–loops, i.e. stem–loops which retain the typical folding structure regardless of the precise location of the start/end of the folded transcript. This is justified by the observation that a pre-miRNA should be robust with respect to the genomic context where it lies. These candidate stem–loops were then scored by an SVM (support vector machine) classifier trained on a set of positive examples derived from known human miRNA precursors and a set of negative examples derived from mRNAs, tRNAs, rRNAs and random regions of the human and viral genomes. The features considered included folding free energy, nucleotide count in the symmetrical stem, and number of A–U, G–C and G:U pairs in the predicted structure. The authors forced the misclassification of positives to be eight times more penalizing than the misclassification of negatives, thus sacrificing sensitivity to higher specificity.
The same approach was then used to predict clustered pre-miRNAs in H. sapiens, M. musculus, and Rattus norvegicus (61) following the observation that many animal miRNAs indeed occur in clusters. The high false positive rate that the approach, in general, could entail is partially mitigated by the fact that only regions close to previously identified miRNAs are scanned, so it is reasonable to assume that these regions are indeed transcribed and can represent instances of clustered miRNAs.
Several other machine learning methods have been proposed to tackle the problem of identifying good miRNA candidates. SVMs have been a popular framework used to learn the distinctive characteristics of miRNAs. Most approaches use sets of features concerning sequence composition (62–64), topological properties of the stem–loop (63–65), thermodynamic stability (63–65), and sometimes other properties including entropy measures (64).
A somewhat different approach called mirCoS-a (66) chains three different SVM classifiers, each focusing on different features of the candidate stem–loops obtained from conserved regions of vertebrate genomes. The aspects covered by each SVM concerned: (i) sequence conservation; (ii) secondary structure conservation; and (iii) location and structure of the mature miRNA in the hairpin structure. By using a chained-filter approach the authors were able to compute complex features for the SVMs downstream in the pipeline, which would have been prohibitively time consuming if applied to all the initial candidates.
An SVM method specifically designed to predict Drosha processing sites is described in (67). The classifier uses 11 features concerning sequence/structure properties in different regions of the stem–loop. This method not only can serve as a pre-processing tool of miRNA candidates as it can also generate additional features for precursor classifiers concerning metrics about the potential processing sites.
Other machine learning methods rely on Random Forests (68) (a method that uses a set of tree-based classifiers combining sampling of training data with random feature selection), a Naïve Bayes classifier (69), or genetic programming (70).
The methods described in this section are natural approaches to the miRNA gene finding problem. The latter is cast as a classification problem and powerful methods are used to generalize from positive and negative examples, as is customary. In this case, however, there are a few questions raised by the positive and negative datasets adopted.
Negative datasets usually include randomly chosen stem–loops extracted from the genome, under the assumption that there is a very low density of pre-miRNAs and therefore there is a small chance of a true miRNA precursor being recruited as a negative example. The number of miRNAs in any given genome is still an open problem and consequently we cannot confidently evaluate the impact of this assumption. Additionally, there may be many stem–loop structures in the genome that would be able to enter the miRNA processing pathway but are not efficiently transcribed, or are simply in the wrong genomic context. Since these machine learning approaches do not usually incorporate any information regarding transcription potential or genomic context, but rather concentrate on stem–loop features, they may be misclassifying an important portion of the search space, despite the fact that cross-validation procedures or validations with independent test sets have given very good measures of sensitivity and specificity.
On the other hand, positive examples are recruited from miRNAs previously identified by experimental procedures or other computational methods and these datasets are, therefore, strongly biased towards highly expressed and extensively conserved miRNAs. This questions the critical assumption that the positive set is truly representative, as low-expression non-conserved miRNAs may have features that are substantially different. Despite this, with a growing number of miRNAs being identified, one can expect an increasingly better performance from these methods.
Target-centered approaches
An innovative strategy to predict miRNA genes is described in (71). The authors aligned the 3′-UTRs of several mammalian genomes and identified highly conserved short motifs showing properties reminiscent of miRNA target seeds. Subsequently, the authors identified hundreds of conserved and stable stem–loops containing conserved sequences complementary to the short motifs previously identified, including several known miRNAs.
Target-centered approaches have the benefit of making few assumptions about the structure of miRNA precursors, but are dependent on the identification of highly conserved motifs in 3′-UTRs which do not represent all the universe of possible targets.
Mixed approaches
Some approaches have combined high-throughput experimental methods with computational procedures in order to identify a wider range of miRNAs. These approaches can use two different strategies: (i) identification of a great number of low-confidence precursor candidates subsequently subject to high-scale experimental verification; (ii) extensive cloning of small RNAs that are then analysed with respect to their localization in the genome and their ability to form stem–loops in the genomic context of the identified locations.
A method called PalGrade (53) followed the former strategy to identify several new conserved and non-conserved miRNAs in H. sapiens. Thousands of candidate stem–loops were selected based on a scoring scheme that considers thermodynamic stability and structural features. The potential expression of this set of candidates was then tested in several tissues with miRNA microarrays, and candidates with strong hybridization signals were further subjected to directed cloning and sequencing. This approach has substantially expanded the catalogue of human miRNAs.
Methods following the second strategy usually consider the bulk of sequenced RNAs, determine their genomic location and apply filters similar to those used by ab initio methods (57,72,73).
As noted before, these approaches cannot, however, detect low-expression or tissue-specific miRNAs. Deep sequencing techniques have formidably expanded our ability to detect low-abundance transcripts but have also presented new challenges. While raising the ability to sequence rare miRNAs, other small transcripts are also amplified and more sophisticated approaches are required to sieve out miRNA transcripts. A method called miRDeep (49) uses a probabilistic model to assess the compatibility of the pattern of sequenced RNA transcripts with properties of miRNA biogenesis. According to this model, a true miRNA precursor should have a characteristic signature, with frequent sequence reads corresponding to the mature region of the stem–loop, and less frequent reads corresponding to other parts of the hairpin structure.
Homology-based searches
Homology-based approaches are a common way of detecting miRNAs that may have been missed by ab initio predictors, and in fact many miRNA gene prediction approaches incorporate an homology-based search as part of their protocol, in addition to the usual search for orthologues which is an integral part of the conservation requirements.
Many homology searches are alignment-based methods and can be applied to the members of the original candidate set that failed to pass some of the filters (50), or specifically directed to regions surrounding known miRNAs in the hope of finding new members of a gene cluster (74). Alternatively, these methods can be used to scan newly sequenced genomes for homologues of known miRNAs (75–77), or to further saturate miRNA gene predictions in previously studied genomes (33).
However, alignment-based methods rely exclusively on sequence conservation. More sensitive methods can be developed by considering structure conservation. An example is the approach described in (78) which proposes a profile-based method using an RNA comparison tool named ERPIN (79) to account for sequence/structure conservation and was able to predict hundreds of new candidates from several different families of animal miRNAs. An alternative example is miRAlign (80).
Another powerful strategy is the use of structure-based clustering. In this approach, a set of candidate structures are clustered using a metric based on sequence/structure alignments. Potential homologues are found in clusters with known miRNAs (81,82).
Computational approaches to miRNA gene finding in plants
Strategies similar to those used in animals have been applied to the prediction of plant miRNA genes. In this case, the problem is considerably more difficult due to the heterogeneous nature of plant pre-miRNA stem–loops which vary greatly in size and structure. Consequently, these methods rely more on the properties of the miRNA:miRNA* duplex within the variable precursor, and it is also not surprising that much fewer approaches have been proposed for plants than for animals.
Filter-based approaches
One of the first methods for identifying miRNAs in plants is described in (83). The authors proposed a workflow that began by identifying all potential hairpins in the intergenic regions of A. thaliana. The hairpins were found by looking for imperfect inverted repeats of 21 nt, representing the putative mature miRNA and corresponding star sequence, that were separated by a distance within a given window. The candidate hairpins were then filtered according to criteria concerning GC content and loop length. The putative miRNA sequences were checked against the rice genome and only those showing high conservation were retained. Finally, the remaining precursor candidates and their orthologues were folded to validate the characteristic stem–loop secondary structure. This procedure suggested 83 new and identified 12 previously known miRNAs. Amongst the miRNA candidates, 19 had their expression experimentally verified, or were found in public databases of small RNAs.
A similar approach is described in (84). The candidate sequences are folded using a secondary structure prediction algorithm and given to a program called MIRcheck. This program receives a sequence/structure specification and the coordinates of a 20-mer within the hairpin and uses a series of metrics concerning the number of unpaired nucleotides and bulges in the miRNA mature regions and the length of the hairpin. Sequences overlapping repetitive elements are eliminated, and a strong conservation criterion is applied retaining only stem–loops where the mature miRNA appears in both genomes and exhibit high conservation in both the miRNA and miRNA* sequence. Additionally, stem–loops are tested for robust folding, indicating that their secondary structures do not change substantially in the presence of flanking sequences. An additional filter consisted in searching for conserved near-perfect complementary matches in the mRNAs of both genomes, presumably target sites for these miRNA candidates. With this method, the authors were able to identify 379 good miRNA candidates in 228 unique loci, of which 23 had their expression experimentally verified.
A computational pipeline called MIRFINDER (85) identifies conserved hairpin structures in the genomes of A. thaliana and Oryza sativa and subsequently applies several filters, based on core features derived from known miRNAs. The features seen in the miRNA reference set suggested that the mature miRNA should be part of a stable continuous helix with no more than a few unpaired or G:U pairs in the miRNA region. The conservation requirements included extensive conservation of the mature miRNA sequence and location in the same stem arm. The authors observed that a large amount of sequences in both genomes could fold into hairpin structures, so a randomization test was setup to assess the statistical significance of the predicted secondary structures. After applying filters for GC content and low complexity sequences, a total of 91 potential miRNA genes were identified, of which 58 had at least one nearly perfect target match.
The methods described so far make extensive use of conservation criteria and are therefore unable to identify miRNAs with less obvious patterns of evolutionary conservation. Other methods have taken advantage of the near-perfect complementarity observed between the miRNA and corresponding target sites in plant mRNAs and were able to identify several novel non-conserved plant miRNAs.
Target-centered approaches
A single-genome approach called findMiRNA (86) replaced the sieve of cross-species conservation of candidate stem–loops with the detection of potential targets within transcripts of the same species. The algorithm starts by indexing all the 7-mers of the intergenic regions, excluding repeats and low GC-content sequences. For each transcript, its overlapping 7-mers are tentatively matched against the index previously computed. For each match, an ungapped alignment of the surrounding areas is produced. The best length-normalized alignment score of size 18–25 is marked as a potential miRNA. If the score is above a given threshold, a dynamic programming algorithm is used to search for a complementary sequence in the vicinity. A secondary structure prediction algorithm is used to verify the presence of a stem–loop structure, and whether the length-normalized MFE is below a given threshold. An additional filter is then used for higher specificity, which exploits the expected typical divergence pattern of miRNA precursors of the same family, whose members have presumably arisen by duplication events. Precursor candidates are put in the same family cluster if they target the same transcript region. Clusters are then scored according to the degree of conservation of the miRNA, miRNA* and intervening sequence, using a scoring function that privileges conservation of the miRNA sequence and penalizes conservation in the intervening region.
A similar approach described in (87), unlike the previous method, does not require that miRNAs be clustered into families. This method takes each mRNA and a genome-wide search is performed in order to identify regions of 20–27 nt that match a portion of the mRNA with at most two mismatches. These matches, termed micromatches, are then used to identify miRNA candidates. The candidates are passed by six filters: (i) high sequence complexity; (ii) no overlap with annotated exons; (iii) no overlap with repeat sequences; (iv) stable miRNA:mRNA duplex; (v) no more than 10 identical copies in the genome; (vi) the putative miRNA is contained in a stable precursor stem-loop structure exhibiting some typical features. An additional sieve is then added that includes only miRNA candidates with more than one target, which is thought to be typical of most plant miRNAs.
Homology-based searches
Upon the identification of an ever increasing number of plant miRNA genes in several species, homology-based search methods begun to be developed seeking the complete enumeration of miRNAs in model organisms (88,89). In general terms, these methods first identify genome hits matching known miRNA mature sequences and then extract the genomic context of such hits and align the candidates with their putative miRNA families followed by the application of some criteria to determine a final list of candidate homologues. More recently, these protocols have been adapted to search for new miRNAs by analysing EST (expressed sequence tag) data (90).
Other approaches
Other methods for plant miRNA gene identification have been developed using a combination of high-throughput sequencing, filtering and machine learning approaches in similar ways to those discussed for animal miRNA prediction (91).
TARGET PREDICTION
The function of a miRNA is ultimately defined by the genes it targets and the effects it has on their expression. Two major silencing mechanisms have been identified for miRNAs: mRNA cleavage, and translational repression. The mechanism of mRNA cleavage is more associated with plants, and seems to be indistinguishable from siRNA-directed cleavage, whereas translational repression is more associated with animals. However, cases of animal miRNAs directing cleavage of mRNAs and plant miRNAs arresting translation are not unknown (33,92–95). More recently, it has been shown that transcript destabilization is common in animals. This down-regulation of the expression of targeted mRNAs has been largely attributed to the sequestration and subsequent degradation of the regulated transcripts onto sub-cellular structures called the cytoplasmic processing bodies (PBs) (40).
Some authors have also suggested a more diverse set of possible action mechanisms for miRNAs (44,96), including a role as RNA guide for mRNA modification, as promoter of DNA methylation, or in the recruitment of specific regulatory proteins (40), amongst other possibilities.
Differences in target complementarity and target location within the mRNA could be related to the silencing mechanism used. Animal targets are preferably located in the 3′-UTR region where the silencing complex can easily interact with the initiation complex and promote attenuation of translation (40). Plant targets, on the other hand, can be located anywhere on the mRNA since a single cleavage site would promote immediate degradation. In addition, the fact that, unlike plants, many animal miRNAs have multiple targets on the same mRNA, and more frequently multiple miRNAs target the same mRNA, may reflect the requirements of the preferred silencing mechanism in metazoa. Other studies suggest that these two silencing mechanisms are determined, for the most part, by the degree of complementarity between a mature miRNA and its target site (20,97), which can have an impact on which coenzymes are recruited to integrate the silencing complex (98).
miRNA targets in animals and plants show important differences and have, therefore, prompted different approaches to their identification. In the remainder of this section we discuss the implications of those differences and the methods proposed to tackle the problem of target prediction in both plants and animals.
Prediction of targets in animals
miRNA targets in animals are usually located in the 3′-UTR region of mRNAs. The latter had already been recognized as an important regulatory region even before the discovery of miRNAs, due to the presence of numerous regulatory signals involved in the control of nuclear export, subcellular localization, transcript stability, amongst other processes (36,99,100). Additionally, this region frequently contains multiple target sites for more than one miRNA (101). It has been shown, however, that target sequences inserted in the coding or 5′-UTR regions can also be functional (37).
Animal miRNA targets are difficult to predict since miRNA:mRNA duplexes often contain several mismatches, gaps and G:U base pairs in many positions, thus limiting the maximum length of contiguous sequences of matched nucleotides (101). However, it is increasingly recognised that near-perfect complementarity between a few bases at the 5′-end of miRNAs and its 3′-UTR targets is instrumental in metazoan target recognition (28,102).
The importance of these seed sites was further reinforced by an experimental study (103) on the principles governing miRNA target recognition. The authors identified two main categories of target sites. The first category are 5′-dominant sites which include canonical sites exhibiting good complementarity at both ends, and seed sites which have poor pairing at the 3′-end, but include a continuous helix of at least 7 bp at the 5′-end. The second category is termed 3′-compensatory sites. These sites exhibit weak pairing at the 5′-end with seeds of 4–6 bp, or seeds of 7–8 bp including bulges, mismatches and G:U pairs which are compensated by strong pairing at the 3′-end. In addition, the authors show evidence suggesting that the most common category is that of seed sites.
In the absence of a clear model for specific miRNA target recognition in animals, most approaches seek conserved 3′-UTR sites with favourable thermodynamic hybridization energies, and use the detection of seed matches as a primary sieve. Other approaches resort to machine learning techniques in an attempt to grasp the rules of target site recognition from the small set of confirmed targets. Despite the success of many of these methods in predicting functional target sites, the number of false positives remains high, particularly for the most sensitive methods (104,105). Two approaches have been proposed to try to achieve better specificity: the use of mRNA expression data, and the incorporation of mRNA secondary structure in the thermodynamic hybridization model.
Seed-based approaches
An early approach (101) searched for targets of Drosophila miRNAs by preparing a database of 3′-UTR sites conserved across two drosophilid species. A distinguishable pattern of better conservation at the region that matches the 5′-end of the miRNA was observed, showing also better complementarity to the miRNA, with few mismatches or G:U pairs in the first 8 nt of that end. This observation prompted the search for conserved sequences that matched the first eight positions of the miRNA. Subsequently, the duplexes thus obtained were ranked by free energy of folding. The statistical significance of the hybridization energies was evaluated against a background of 10 000 randomly selected target sites. Several instances of multiple target sites per mRNA were observed and while single hits were generally not statistically significant, the combined score of the binding sites per mRNA generally was, which led to the idea that several regulatory sites were required for efficient regulation. This approach predicted several targets, including five previously validated sites. Three targets from the novel predictions were experimentally verified.
A method called MiRanda (106) considers all the known miRNAs of D. melanogaster. The algorithm encompasses three phases. In the first phase, the miRNAs are matched against the 3′-UTR regions of all possible targets allowing for G:U pairs as well as indels of moderate size. The method does not rely on seed matches directly but privileges complementarity at the 5′-end of the miRNA by using a scaling factor for scores computed in this region, and incorporates some position-specific empirical rules. The second phase consists in computing the thermodynamic stability of the miRNA:target duplex, and the third and final phase is an assessment of the evolutionary conservation of miRNA–target associations across two additional species. Finally, using a randomization procedure, the authors estimated the false positive rate and showed that it is reduced if one considers only mRNAs with multiple target sites. The same approach was later used to predict targets in humans and other vertebrates (107).
The first method to explicitly use the concept of seed matches was TargetScan (108). The algorithm takes miRNAs conserved across a group of organisms and scans a set of orthologous 3′-UTR sequences from these organisms. Seed matches are defined as small segments of 7 nt that ought to have perfect complementarity to the bases in positions 2–8 of the miRNA. These matches are then extended to target sites involving the entire miRNA, allowing for G:U pairs, and using a folding algorithm to predict the secondary structure of the heteroduplex. To each putative target, a folding free energy value is assigned, and a Z-score is calculated based on the number of matches predicted in the same target transcript and respective free energies. The candidate transcripts for each organism are ranked by Z-score, and the process is repeated for each organism in the set. Cut-off values for rank and Z-score are given, and the final candidate set is composed of targets that respect the established limits for all orthologous transcripts.
The same authors would later add more organisms to their working set (109), which enabled them to relax both the rank and score cut-offs and rely exclusively on seed matches consisting of a segment of only 6 nt while still improving the signal/noise ratio. More improvements were attained by analysing the sequences flanking the 6-nt seeds, which would show a bias towards the presence of certain nucleotides in key positions, particularly an adenosine at the 3′-end of the target site.
An independent study (109) examined a set of known miRNA binding sites in C. elegans, and identified a pattern of consecutive GC-rich base-pairings with the miRNA that was termed binding nucleus. A scoring scheme for the binding nucleus was devised by considering the weighed sum of consecutive GC, AU and G:U pairs. The weights were determined by computing the values that maximized the difference of the mean scores of the known binding sites and the scores obtained for random sequences generated using site-independent background frequencies derived from C. elegans 3′-UTR regions, divided by the standard deviation of the background scores. A threshold score was defined for each miRNA by observing the score distribution over a random sequence thus allowing the computation of a P-value. Interestingly, the authors observed that the nucleus was typically 6–8 nt long and located near the 5′-end of the miRNA.
The tool DIANA-microT (11) was used to predict new miRNA targets in H. sapiens. The authors searched for targets of 10 miRNAs which were conserved in M. musculus in the set of all repeats-masked human 3′-UTR sequences. The search method considered two hypotheses about miRNA:mRNA regulatory associations: (i) they should be conserved high-affinity interactions; (ii) they should be structurally restrained due to the enzymology of the miRISC complex. The first observation resulted in an algorithm to compute the thermodynamic stability of imperfect miRNA:mRNA pairings. The second hypothesis led to the speculation that the structural restraints might be reduced to a set of general rules. In order to identify these rules, the authors performed a series of experiments whereby some putative target site sequences were cloned onto a reporter construct. These rules were then used to filter the initial set of candidates. The results obtained with these experiments once again underlined the importance of near-perfect complementarity on the first few nucleotides at the 5′-end of the miRNA.
The first tool that could be used on a single-genome was RNA-hybrid (112). This method consists in a dynamic programming algorithm that calculates the energetically most favourable hybridization of a miRNA and its target mRNA, while also allowing the user to specify a portion of the miRNA that should form a perfect helix, corresponding to the seed site. The statistical significance of the predicted targets is determined using extreme value statistics for minimum free energies normalized for target length, and a Poisson distribution is used to model multiple binding sites of a miRNA for the same target. The statistical treatment is extended with a comparative analysis of conserved binding sites in orthologous targets of related species.
A popular tool, called PicTar (113), is a combinatorial method that identifies individual miRNA target sites by searching near-perfect seeds defined as a stretch of ∼7-nt starting at position 1 or 2 from the 5′-end of the miRNA. These target sites are then filtered with respect to the MFE of the heteroduplexes and to whether these sites fall into overlapping positions across the aligned orthologous sequences. The target sites that pass both these filters are termed anchors. Sequences that show a user-defined minimum number of anchors are then ranked using an HMM maximum likelihood score. This score is computed considering all segmentations of the target sequence into target sites and background, thus accounting for the synergistic effect of multiple binding sites for a single miRNA or several miRNAs co-regulating the same transcript.
The MovingTargets tool (114), relies on a database of potential miRNA targets obtained through the identification of highly conserved segments of not more than 50 nt on orthologous 3′-UTR regions of two close species. Target sites for a set of given miRNAs are sought on this database according to five user-adjustable criteria: (i) number of target sites in the mRNA; (ii) stability of the miRNA:mRNA hybridization, as measured by the MFE; (iii) number of consecutive base pairs in the heteroduplex involving the 5′-end of the miRNA; (iv) total number of paired nucleotides in the 5′-end of miRNA; (v) number of G:U base pairs in the 5′-region of the miRNA.
A later approach was entirely based on network-level conservation of seed matches (115). The method begins by exhaustively enumerating all the k-mers of lengths 7, 8 and 9 which are conserved across orthologous 3′-UTRs of worm and fly genes. A conservation score measures the overlap between the sets of orthologous regions containing at least one copy of a given k-mer, and these scores are then compared with those obtained with a control assay done over randomized 3′-UTRs. The results show that high scoring k-mers score much higher in the real data than in the control. Some of these k-mers are known to be involved in post-transcriptional regulation and many of them are complementary to the 5′-ends of known miRNAs, more often than what would be expected by chance. Most high-scoring k-mers identified in worms were also conserved in flies and vice-versa. Candidate targets were identified as those genes whose 3′-UTR and that of its orthologue contain a high-scoring k-mer which is also complementary to the 5′-end of a miRNA.
The first method to seek to distinguish between sites that are conserved from those which are under selective pressure (116) used a Bayesian framework providing a probabilistic model to assess whether a given conserved site was under evolutionary selection. The method relies on pairwise alignments of several orthologous 3′-UTRs. Seed matches exhibit a particular conservation pattern and the method sets out to compute the probability of that site being under selection in at least one other species than the reference, given the observed conservation pattern.
Machine learning approaches
TargetBoost (117) is a machine-learning method that combines genetic programming with boosting. Instead of relying on criteria based on sequence complementarity, thermodynamic stability or evolutionary conservation, TargetBoost tries to learn the hidden rules of miRNA:target site hybridization. The genetic programming component consists in spawning and evolving a series of pattern sequences which try do describe the general properties of miRNA target sites, namely the existence of a nucleus of consecutive paired bases (seed) or a bulge of unpaired nucleotides. Each of these pattern sequences is a classifier, and they are all combined using the boosting technique that gives each classifier a weight depending on its performance on the training set. Additional filters can be added to this procedure, like the verification of evolutionary conservation or the existence of multiple target sites in the same 3′-UTR.
Other machine learning approaches using the popular SVM framework have been proposed. These approaches try to generalize from a modest set of experimentally verified positive and negative examples. An example is miTarget (118) which uses an SVM considering structural features of the 5′ and 3′ half of the hybridization site, thermodynamic features and positional features.
Integration of target gene expression data
Initially it was thought that animal miRNAs, by interacting with multiple sites on 3′-UTRs, would inhibit the accumulation of protein products of the targeted messages without affecting the level of expression of the corresponding mRNAs (28). However, it is now clear that, in many cases, there is a direct impact on the concentration of mRNA transcripts (39). However, there are still many documented miRNAs which have no impact on target mRNA levels, whose influence cannot explain by itself the observed decrease in protein accumulation, or which are, more plausibly, independently down-regulated at the level of transcription (40,119).
Nevertheless, several target prediction methods incorporating putative target expression levels have been developed (120–122) and have proved to be a valuable approach to target identification.
Integration of target secondary structure
Some authors tried early to implicitly incorporate target secondary structure as a measure of site accessibility in their prediction methods (123,124). However, a major progress in the understanding of miRNA:target recognition mechanisms was made with the development of a thermodynamic model that incorporates measures of accessibility of target sites (125). According to this model, a crucial determinant of effective binding is the change of free energy between the unbound 3′-UTR, with its pre-existing secondary structure, and the hybridized state. This model permitted the development of a new target prediction method called PITA.
More recently, another method (126) takes advantage of a large dataset of experimentally verified miRNA:mRNA associations to derive a scoring scheme that combines site conservation, 5′-seed pairing, structural accessibility and hybridization energy criteria, illustrating the need to combine several features in order to accurately identify new targets.
Prediction of targets in plants
Unlike the case with animals, plant miRNAs generally show a near-perfect complementarity with their targets on mRNAs (29,44) which immensely facilitates computational searches.
Taking advantage of this recognized property, a method (44) was developed to search for antisense hits of known miRNAs on Arabidopsis mRNAs. The matches were required to have no gaps and only canonical pairs. The same search was performed on a randomized version of the sequences with identical size and base composition, a procedure which validated the statistical significance of the obtained results. Of the 16 miRNAs used in this search, 14 had targets with less than four mismatches. The authors applied the same procedure to C. elegans and D. melanogaster and found that the number of hits obtained was not significantly different from the number of hits seen with the randomized version of the sequences, which suggests that, although some animal targets may exhibit near-perfect complementarity, these do not represent the general case. A very similar approach is described in (83). Later, it was observed that a more sensitive method could be obtained by being more lenient with the quality of the miRNA:mRNA pairing and adding a requirement for conservation with respect to a close species suggesting that at least some pairings could be less perfect than anticipated (84).
CONCLUSION AND PERSPECTIVES
Most computational approaches developed so far make extensive use of evolutionary conservation either to predict miRNA genes or miRNA:target associations. This illustrates our collective ignorance of the subtle rules presiding miRNA biogenesis and target specificity. Since the cell cannot use the filter of evolutionary conservation (28) to choose among all potential stem–loops or all putative targets, we seem to be missing a significant part of the whole story. Future developments ought to focus on the need to establish more accurate models for these central problems.
The search for the distinguishing characteristics of animal miRNA precursors continues. For the known miRNAs, it has been already established that they have particularly low-energy structures compared with other RNAs, even when corrected for size and GC content (127,128). Additionally, these stem–loops seem to be robust with respect to their genomic contexts, as should be expected for efficient Drosha recognition. They are also stable in the sense that a similar base-pairing pattern persists for a set of sub-optimal structures making up the thermodynamic ensemble where the stem–loop should be found most of the time. Mutational robustness is another property that has been suggested for miRNA precursors (129), but it is most likely observed in ancient well-conserved pre-miRNAs rather than more recent non-conserved precursor stem–loops.
A better insight for future developments on miRNA gene finding can probably be attained by considering what is not a miRNA. A given location in the genome does not contain a miRNA if: (i) it is not efficiently transcribed; (ii) it does not contain a stem–loop structure amenable to efficient processing by all participants in the miRNA biogenesis pathway; (iii) it cannot regulate a target gene in a physiologically relevant manner. Most available methods have focused on the second item and little attention has been paid to the other two.
Another question that becomes more important when many authors argue that the identification of well-conserved and phylogenetically extensive miRNAs is reaching its saturation is whether non-conserved, presumably more exotic, miRNA precursors would be processed as such in different organisms that may have small yet important differences in their processing pathways. The elucidation of this question is crucial to methods which try to generalize from pre-miRNAs taken from several different species.
The area of target prediction has received a new impetus with the recent proposal of a thermodynamic model incorporating target accessibility. However, seed matches constitute an important sieve to control false positives. The seed hypothesis, adopted almost unanimously by current target prediction methods, was recently reinforced with a study that obtained the structure of an important component of the silencing complex bound to a DNA guide-strand, and which lays down the biochemical basis for the role of seed sites (130). However, at least some experimentally confirmed targets seem to violate the seed rule by including mismatches or G:U pairs (109,131). The present scarcity of confidently validated miRNA targets, establishing not only miRNA–target associations, but specifically pinpointing the hybridization sites, is the greatest obstacle not only to the development of better prediction methods but also to the systematic assessment of the performance of current tools.
Target prediction becomes even more challenging with the discovery that RNA editing is common in miRNAs (31,132,133). This could substantially change the mature sequence and, consequently, the specificity of its targets. Moreover, a study conducted on human miRNA targets (134) shows that miRNAs tend to target genes with distinctively AT-rich 3′-UTR regions, even when these genes are located in GC-rich isochores, suggesting an unknown function for this compositional bias. The authors argue that better knowledge of the background distribution of nucleotides in 3′-UTR regions may lead to improvements in miRNA target predictions.
For the future, we need a better understanding of the biochemical requirements for Drosha and Dicer processing. We also need to integrate promoter evidence in miRNA gene prediction algorithms, as well as evolutionary models for pre-miRNAs and 3′-UTRs if we are to continue to use comparative approaches for miRNA gene and target predictions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FCT (Portuguese Science Foundation), PDCT; ERA-NET programs (projects PTDC/EIA/67722/2006 and ERAPGFP/06.030A) PhD grant (SFRH/BD/29246/2006 to N.D.M.); the ANR (REGLIS project NT05-3_45205); INRIA (one-year stay of A.T.F. in Lyon, partial). Funding for open access charge: Fundação para a Ciência e a Tecnologia, (project PTDC/EIA/67722/2006).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 4.Lai EC. microRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baskerville S, Bartel DP. Microarray proling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 16.Cullen BR. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 20.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J. Biol. Chem. 2004;279:42230–42239. doi: 10.1074/jbc.M404931200. [DOI] [PubMed] [Google Scholar]
- 25.Lin S-L, Chang D, Ying S-Y. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorodkin J, Havgaard JH, Ensterö M, Sawera M, Jensen P, Ohman M, Fredholm M. MicroRNA sequence motifs reveal asymmetry between the stem arms. Comput. Biol. Chem. 2006;30:249–254. doi: 10.1016/j.compbiolchem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Bartel B, Bartel DP. MicroRNAs: at the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 31.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNAprecursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying S-Y, Lin S-L. Intronic microRNAs. Biochem. Biophys. Res. Commun. 2005;326:515–520. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- 33.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 34.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y-K, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 37.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RHA. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 40.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 43.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 45.Sunkar R, Zhu J-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millar AA, Waterhouse PM. Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics. 2005;5:129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 47.Chitwood DH, Timmermans MCP. Target mimics modulate miRNAs. Nat. Genet. 2007;39:935–936. doi: 10.1038/ng0807-935. [DOI] [PubMed] [Google Scholar]
- 48.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, vanDyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 49.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 50.Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, Ruvkun G, Kim J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 51.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 54.Berezikov E, Guryev V, van deBelt J, Wienholds E, Plasterk RHA, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 55.Ohler U, Yekta S, Lim LP, Bartel DP, Burge CB. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 57.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandmann T, Cohen SM. Identification of novel Drosophila melanogaster MicroRNAs. PLoS ONE. 2007;2:e1265. doi: 10.1371/journal.pone.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose D, Hackermueller J, Washietl S, Reiche K, Hertel J, Findeiss S, Stadler P, Prohaska S. Computational RNomics of drosophilids. BMC Genomics. 2007;8:406. doi: 10.1186/1471-2164-8-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nam J-W, Shin K-R, Han J, Lee Y, Kim VN, Zhang B-T. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, vanNimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue C, Li F, He T, Liu G-P, Li Y, Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hertel J, Stadler PF. Hairpins in a haystack: recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–e202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
- 64.Ng KLS, Mishra SK. De novo SVM classification of precursor microRNAs from genomic pseudo hairpins using global and intrinsic folding measures. Bioinformatics. 2007;23:1321–1330. doi: 10.1093/bioinformatics/btm026. [DOI] [PubMed] [Google Scholar]
- 65.Huang T, Fan B, Rothschild M, Hu Z, Li K, Zhao S. MiRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics. 2007;8:341. doi: 10.1186/1471-2105-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheng Y, Engström PG, Lenhard B. Mammalian MicroRNA prediction through a support vector machine model of sequence and structure. PLoS ONE. 2007;2:e946. doi: 10.1371/journal.pone.0000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helvik SA, Snøve O, Saetrom P. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics. 2007;23:142–149. doi: 10.1093/bioinformatics/btl570. [DOI] [PubMed] [Google Scholar]
- 68.Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yousef M, Nebozhyn M, Shatkay H, Kanterakis S, Showe LC, Showe MK. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics. 2006;22:1325–1334. doi: 10.1093/bioinformatics/btl094. [DOI] [PubMed] [Google Scholar]
- 70.Brameier M, Wiuf C. Ab initio identification of human microRNAs based on structure motifs. BMC Bioinformatics. 2007;8:478. doi: 10.1186/1471-2105-8-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 73.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatterjee R, Chaudhuri K. An approach for the identification of microRNA with an application to Anopheles gambiae. Acta Biochim. Pol. 2006;53:303–309. [PubMed] [Google Scholar]
- 76.Weaver D, Anzola J, Evans J, Reid J, Reese J, Childs K, Zdobnov E, Samanta M, Miller J, Elsik C. Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biol. 2007;8:R97. doi: 10.1186/gb-2007-8-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Norden-Krichmar T, Holtz J, Pasquinelli A, Gaasterland T. Computational prediction and experimental validation of Ciona intestinalis microRNA genes. BMC Genomics. 2007;8:445. doi: 10.1186/1471-2164-8-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Legendre M, Lambert A, Gautheret D. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics. 2005;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- 79.Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 81.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ritchie W, Legendre M, Gautheret D. RNA stem-loops: to be or not to be cleaved by RNAse III. RNA. 2007;13:457–462. doi: 10.1261/rna.366507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X-J, Reyes JL, Chua N-H, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 85.Bonnet E, Wuyts J, Rouzé P, dePeer YV. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl Acad. Sci. USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lindow M, Krogh A. Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics. 2005;6:119. doi: 10.1186/1471-2164-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y, Li W, Jin Y-X. Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim. Biophys. Sin. 2005;37:75–87. [PubMed] [Google Scholar]
- 89.Dezulian T, Remmert M, Palatnik JF, Weigel D, Huson DH. Identification of plant microRNA homologs. Bioinformatics. 2006;22:359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- 90.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 91.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu J. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 93.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 96.Sontheimer EJ, Carthew RW. Silence from within: endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 97.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 99.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat. Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 100.Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 101.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 105.Mazière P, Enright AJ. Prediction of microRNA targets. Drug. Discov. Today. 2007;12:452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 106.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lewis BP, Shih I-H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 109.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 110.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 111.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, daPiedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 114.Burgler C, Macdonald PM. Prediction and verification of microRNA targets by MovingTargets, a highly adaptable prediction method. BMC Genomics. 2005;6:88. doi: 10.1186/1471-2164-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan CS, Elemento O, Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaidatzis D, vanNimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saetrom O, Snøve O, Saetrom P. Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA. 2005;11:995–1003. doi: 10.1261/rna.7290705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim S-K, Nam J-W, Rhee J-K, Lee W-J, Zhang B-T. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 120.Wang X, Naqa IME. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 121.Huang JC, Frey BJ, Morris QD. Comparing sequence and expression for predicting microRNA targets using GenMiR3. Pac. Symp. on Biocomput. 2008:52–63. doi: 10.1142/9789812776136_0007. [DOI] [PubMed] [Google Scholar]
- 122.Cheng C, Li LM. Inferring microRNA activities by combining gene expression with microRNA target prediction. PLoS ONE. 2008;3:e1989. doi: 10.1371/journal.pone.0001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc. Natl Acad. Sci. USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thadani R, Tammi MT. MicroTar: predicting microRNA targets from RNA duplexes. BMC Bioinformatics. 2006;7(Suppl. 5):S20. doi: 10.1186/1471-2105-7-S5-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 126.Hammell M, Long D, Zhang L, Lee A, Carmack C, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008 doi: 10.1038/nmeth.1247. doi:10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bonnet E, Wuyts J, Rouzé P, dePeer YV. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 128.Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. 2006;63:246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Borenstein E, Ruppin E. Direct evolution of genetic robustness in microRNA. Proc. Natl Acad. Sci. USA. 2006;103:6593–6598. doi: 10.1073/pnas.0510600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem. Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 132.Blow MJ, Grocock RJ, vanDongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–1176. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 134.Robins H, Press WH. Human microRNAs target a functionally distinct population of genes with AT-rich 3′ UTRs. Proc. Natl Acad. Sci. USA. 2005;102:15557–15562. doi: 10.1073/pnas.0507443102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.