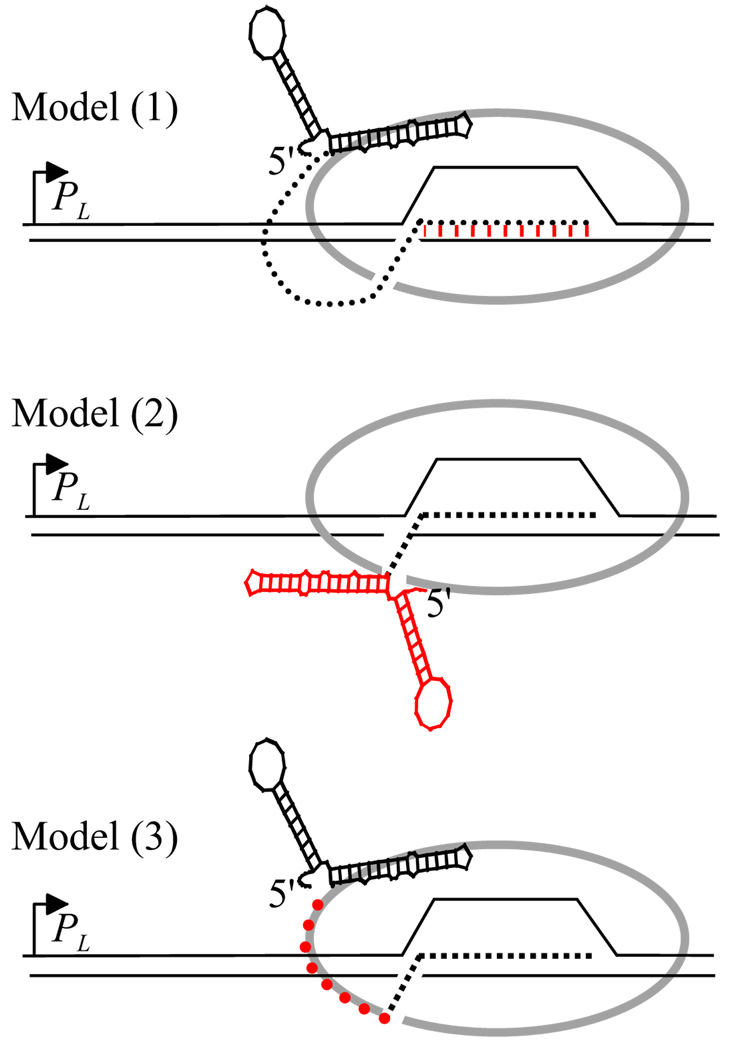
Fig. 4.
Models of put-mediated pause suppression. Each diagram shows RNAP (grey ovals) that has initiated at the HK022 PL promoter, translocated through the putL site and stopped at the U-rich pause. The end of the RNA exit channel is indicated by a break in the grey oval, RNA by dotted lines (except for put) and DNA by thin lines. In model (1), putL binds to and modifies RNAP. The modification suppresses backtracking and termination by the same mechanism. For purposes of illustration we show the modification as an alteration in the RNA:DNA hybrid (red hatching), although in fact the nature of the modification is unknown (see text). In model (2), the secondary structure of putL (in red) prevents the re-entry of RNA that is immediately upstream of the EC into the exit channel. Binding of putL RNA to RNAP is needed to suppress termination but not pausing. In model (3), putL RNA binds close to the exit channel as or shortly after it emerges from the enzyme, and the short size of the polyribonucleotide chain between the end of anchored putL RNA and the end of the channel (in red) prevents RNA re-entry. Further elongation of the transcript relieves this restriction. The bound putL RNA suppresses termination by an unknown mechanism.