Figure 1.
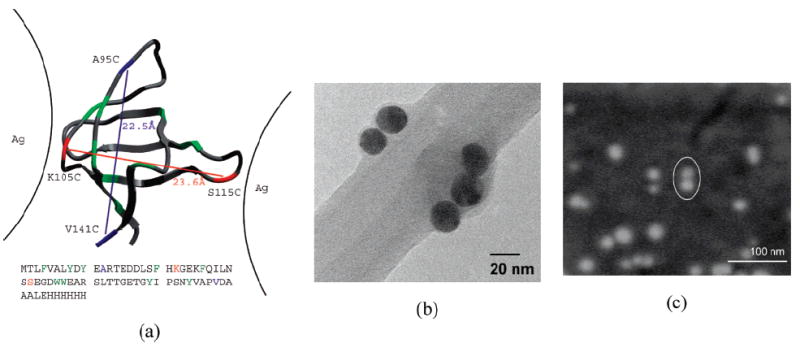
(a) Two small FynSH3 proteins used as a linker between Ag-NPs (denoted K105C/S115C and A95C/V141C). The protein is 70 amino acids long, but only 56 are structured, as shown. The remaining unstructured portion of the protein is represented by a methyl group at the beginning and 13 amino acids His-tag on the end. The cysteine sites bearing sulfur atoms are indicated and connected by lines that denote pairs present in individual constructs. The native-state distance between the two disulfide groups and the sequence separation of each construct are also shown. (b) TEM image of a small NPs-dimer and trimer resulting from cross-linking with the K105C/S115C mutant. The Ag-NPs can be identified as black, round particles that adhere to the inner edge of the holes in the lacey carbon film of the grid. (c) SEM images of a small NPs dimer (marked by a white oval) resulting from cross-linking with K105C/S115C. The NPs dimer appears in this case as two very close white dots. At such low concentrations (i.e., 5 × 10−9 M), most of the examined spots on the silicon wafer contain monomers, and only a small percentage of the observed particles is represented by dimers or other small aggregates.
