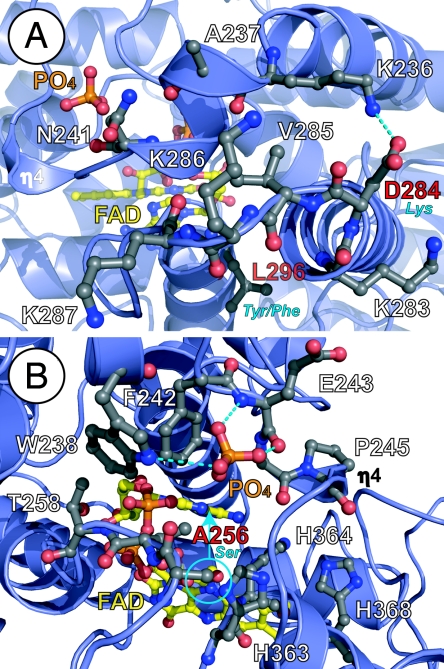
Fig. 4.
Environmental tuning of FAD for diverse functions of the 6-4/clock cluster. (A) The phosphate-binding motif (top) with bound phosphate (orange) and PKL protrusion motif (bottom) partially block substrate access (from left) to FAD (yellow). The PKL protrusion motif varies in sequence at CRY NLS (KVRK/R) equivalent to At64PHR 284-DVKK-287 (labeled in white, with sequence difference in red/blue) and the hydrophobic switch (bottom center), where At64PHR Leu-296 (red label) becomes aromatic (blue label), to distinguish CRY1(Tyr) from CRY2(Phe). (B) Conserved phosphate-binding motif (Asp-235–Ala-256) in 6-4/clock cluster encircles and hydrogen bonds (dashed lines) to phosphate anion (orange) in At64PHR, but can likely bind phosphorylated serine (blue circle) of clock CRYs (equivalent to At64PHR Ala-256, red label). Conformational changes in this motif may impact substrate access to FAD (from right) and binding near the signature His–His–Leu–Ala–Arg-His motif (lower right, with His residues labeled) required for catalysis. Helices and loops are shown with blue ribbons and loops, respectively, and pertinent side chain with ball-and-stick models, are colored by atom type.