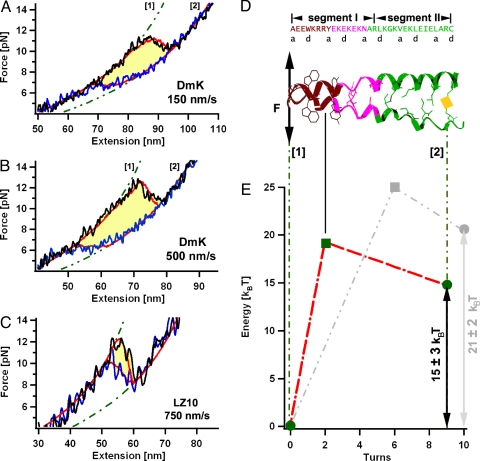
Fig. 2.
Mechanical unzipping profiles and mapped energy landscapes of the kinesin neck (DmK) and a leucine zipper. (A and B) Averaged forward (black) and backward (blue) force traces pulled at 150 nm/s (A) and 500 nm/s (B) measured on the DmK coiled coil. Red lines are the results of the kinetic Monte Carlo simulation. Green dotted lines represent the WLC elasticity (19) for polypeptides with fixed contour length. (C) For comparison, the averaged force traces from a canonical coiled coil (LZ10) at velocities of 750 nm/s are shown. (D) Sequence of the DmK-neck coiled coil aligned to the structure model of the comparable neck coiled coil from Rattus norwegicus (13). The C-terminal half (segment II) shows the canonical knob-into-holes scheme, whereas within segment I noncanonical amino acids are found. (E) Energy landscape of the DmK coiled coil (red) and the LZ10 coiled coil (gray) used for the Monte Carlo simulation.