Figure 9.
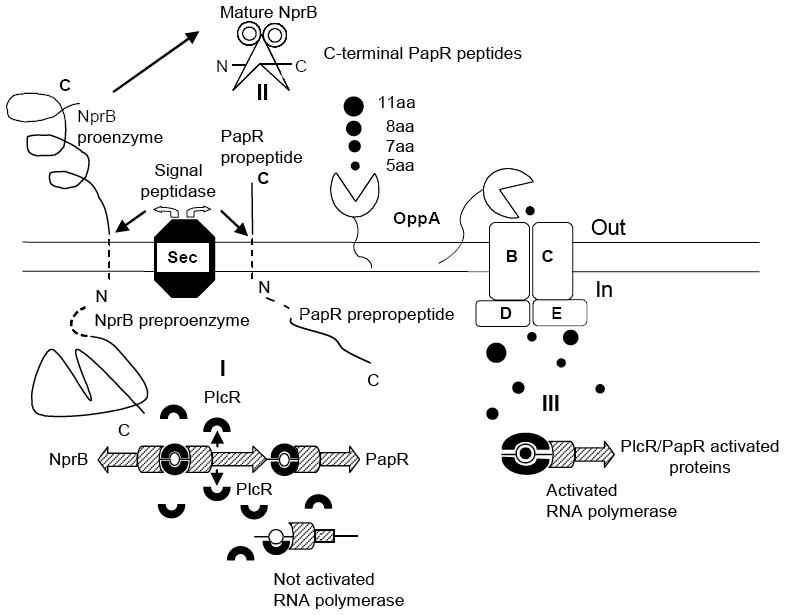
Model of PlcR/PapR regulon activation in B. cereus 569 strain. (I) Schematic diagram of intracellular synthesis of PlcR (indicated by horseshoes), NprB and PapR preproenzymes (both indicated by line figures with dashed N-terminal sequences). The plcR, papR and nprB transcription is initiated by RNA polymerases (indicated by hatched figures) activated by only a PlcR dimer bound to the recognition sites. (II) Extracellular NprB processing of pro-PapR into a set of C-terminal peptides with the consequent transport of the peptides into the cell via oligopeptide permease complex Opp ABCDE. (III) Intracellular assembly of the transcriptional complex including PlcR protein with corresponding C-terminal peptide, PlcR-binding site and RNA polymerase. RNA polymerase is activated when the C-terminal peptide is bound to PlcR and the corresponding PlcR-binding site that is located in the vicinity of the RNA polymerase sigma subunit recognition site. Without the corresponding C-terminal peptide the initiating transcriptional complex is not assembled and RNA polymerase is not activated as illustrated in region I. See text for details.
