Abstract
Aquaporin (AQP) 4 is the predominant water channel in the mammalian brain, abundantly expressed in the blood–brain and brain–cerebrospinal fluid interfaces of glial cells. Its function in cerebral water balance has implications in neuropathological disorders, including brain edema, stroke, and head injuries. The 1.8-Å crystal structure reveals the molecular basis for the water selectivity of the channel. Unlike the case in the structures of water-selective AQPs AqpZ and AQP1, the asparagines of the 2 Asn-Pro-Ala motifs do not hydrogen bond to the same water molecule; instead, they bond to 2 different water molecules in the center of the channel. Molecular dynamics simulations were performed to ask how this observation bears on the proposed mechanisms for how AQPs remain totally insulating to any proton conductance while maintaining a single file of hydrogen bonded water molecules throughout the channel.
Keywords: brain edema, inhibitor discovery, NPA motif
The aquaporin (AQP) family includes both AQPs that conduct water, but not glycerol, and aquaglyceroporins that mediate diffusion of water, glycerol, and certain other small molecules in their neutral form across biological membranes. In humans, 13 different AQPs (AQP0–12) provide for transport in different tissues, each of which has broad clinical importance (1, 2). Besides AQP4, AQP1 and AQP9 are also expressed in the brain (3); AQP1 is expressed in the epithelial cells of the choroid plexus, and has a role in cerebrospinal fluid production, whereas AQP4 is localized to the endfeet of astrocytes in contact with the blood vessels of the blood–brain barrier and in astrocytic processes in contact with synapses. From its tissue-specific concentrated localization in closely packed tetragonal arrays, and the improved response to water intoxication or stroke in AQP4−/− knockout mouse, it is thought that AQP4 is primarily responsible for cerebral water homeostasis (4). AQP4 also may be involved in buffering altered potassium ion concentration after neuronal activity due to its codistribution with KIR4.1 potassium channels in synapses (5). AQP9 is an aquaglyceroporin also found in astrocytes, and with suggested roles in glycerol and monocarboxylate diffusion and energy metabolism in catecholaminergic neurons.
The secretion and absorption of cerebrospinal fluid is precisely controlled, because the brain is encased within the rigid cranium; thus, any increase in intracranial pressure caused by edema can lead to compression of brain tissues resulting in neurological disorders and cell death. Because of its role, inhibitors of AQP4 are sought, although so far with conflicting results. Certain quaternary ammonium compounds (6), antiepileptic drugs (7, 8), and serotonin receptor agonists (9) have been reported to inhibit AQP4 water transport in oocytes with IC50 values down to the low micromolar range. However, Yang et al. (10) report no inhibition up to 100 μM using different assay methods. Therefore, the atomic resolution structure of human AQP4 provides the means of discovering and validating therapeutic agents that might diminish damage from stroke, tumor-associated edema, epilepsy, traumatic head injury, and other CNS disorders associated with brain water imbalance.
AQP4 is the primary target in the autoimmune disease neuromyelitis optica (NMO). Primarily affecting the optic nerves and spinal cord, AQP4-specific autoantibodies (NMO-IgG) activate the complement-mediated inflammatory demyelination and necrosis (11). Knowing the structure around the epitopes for the NMO-IgG can facilitate discovery of agents that may compete for, or alter the site without triggering the complement cascade.
On a structural level, AQP4 is unique among AQPs that it exists in 2 isoforms owing to the use of 2 different translation initiation sites at methionine M1, or at M23. The M1 and M23 isoforms have very different effects on array formation with the shorter isoform favoring larger arrays mediated by 2 symmetric interactions between Arg-108 of each molecule and Tyr-250 of another molecule in the neighboring tetramer (12, 13). The C-terminal 3 amino acids, −SSV, serve as the ligand of a PDZ binding partner, α-syntrophin, which is a component of the dystrophin protein complex that links AQP4 to the actin cytoskeleton (14). Such bridged connection between AQP4 and the actin cytoskeleton allows AQP4 to be anchored at the endfeet of astrocytes such that transgenic mice deficient in α-syntrophin completely lack such polarized expression in astrocytes (15).
AQP4 is not sensitive to inhibition by mercury (16), because it does not have the reactive cysteine residue in the lumen of the channel corresponding to Cys-191 in AQP1 (17). AQP4 conductance is reduced >50% by phosphorylation mediated by protein kinase C at Ser-180 (18, 19), and increased ≈40% by protein kinase G activity at Ser-111 (20). The gating mechanism by phosphorylation events may be similar to that of the spinach AQP SoPIP2;1 (21, 22).
To delineate the cellular organization, immune related properties, and the prospects for inhibitor development, we determined the 1.8-Å crystal structure of human AQP4 from heterologously expressed protein. To date, there are only 3 crystal structures reported for mammalian AQPs, 2 purified from naturally rich sources, AQP1 in red blood cells, AQP0 from the eye lens (23, 24), and human AQP5, from protein heterologously expressed in Pichia pastoris (25).
Results
Overall Architecture.
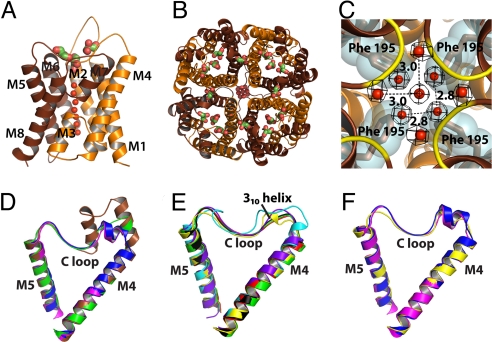
Crystals of the M1 isoform full-length AQP4 diffracted to ≈8 Å in space group I4. Trypsinization improved the resolution. Both full-length and trypsinized protein (hAQP4) have water conductance in reconstituted proteoliposomes (Fig. S1a). The hAQP4 crystallized in space group P4212, although packing interactions between the tetramers are completely different from in the electron diffraction structure of rAQP4 (13). The rAQP4 structure was determined to 3.2 × 3.6 Å with no water or glycerol molecules observed. The X-ray structure of hAQP4 at 1.8-Å resolution shows water molecules throughout the channel, 5 glycerol, and 1 octyl glucoside molecule. Each monomer, surrounded by 6 and 2 half-length alpha-helices (M1 to M8), tetramerizes along the crystallographic 4-fold c axis (Fig. 1 A and B) (26).
Fig. 1.
General features. (A and B) Monomer and tetramer views of hAQP4 in diagram representation. Brown and orange colors represent the N- and C-terminal pseudo 2-fold related portions. Water molecules are represented as red spheres, and glycerol molecules are shown as green sticks. (A) The side view of the monomer. Helices are labeled M1 to M8. (B) The tetramer viewed from the extracellular side down the crystallographic 4-fold symmetry axis. (C) The network of water molecules found at the intracellular side of the central pore. The central pore is at the crystallographic 4-fold symmetry axis, and is formed by the tetramer. The 2Fo-Fc density of the water molecules is shown in black, contoured at 1.2 σ. The backbone amides of Ser-188 and Gly-189 are colored yellow in diagram representation. Phe-195 is shown as brown stick and cyan surface. (D–F) Diagram representation of the C loop of all of the AQP X-ray structures solved to date. (D) E. coli GlpF (brown), archeal AqpM (magenta), spinach AQP SoPIP2;1 (blue), and PfAQP (green). (E) Rat AQP4 (yellow), human AQP4 (black), human AQP5 (red), E. coli AqpZ (cyan), bovine AQP0 (green), and bovine AQP1 (purple). (F) Comparison of the 310 helix of rat AQP4 (yellow) with the 2-turn helix of AqpM (magenta) and spinach AQP (blue). All structural renderings were made with PyMOL (http://www.pymol.org).
Central 4-Fold Axis.
The physiological 4-fold axis insulates against all solutes and water. On the cytoplasmic side, a 4-fold arrangement of water molecules is stabilized by the backbone amides of Ser-188 and Gly-189 (Fig. 1C). Throughout ≈21 Å of the midmembrane section, Phe-195, Leu-191, and Leu-75, repeated 4 times, create a hydrophobic block. This observation contrasts with the 4-fold axis in Plasmodium falciparum aquaglyceroporin, PfAQP, where the region is blocked by 4 aliphatic chains of phospholipids or fatty acids (27), and the human AQP5 where a single lipid molecule is found (25).
Rat AQP4 and Human AQP4.
Although hAQP4 and the electron diffraction structure of rAQP4 in lipid bilayers crystallize in the same space group (P4212), their crystal lattice contacts lie on different surfaces of the protein. The hAQP4 3D crystal contains head-to-head contacts only, because tetramers within the horizontal plane are too far apart (a = 82.1 Å) to make contact with each other (Fig. S2a). The rAQP4 2D crystal lattice has tetramers closer together (a = 69.0 Å), and contains both in-plane and between-plane contacts of the latticed tetramers. Based on the molecular contacts in the crystal, the interaction between the short 310 helices in the C loop was proposed to be a possible mechanism for AQP4-mediated cell–cell adhesion (Fig. S2b) (13). Although the sequences of the C loop are the same in hAQP4 and rAQP4 (Fig. S3), hAQP4 does not adopt the short 310 helix in this region (Fig. S4).
Extracellular Vestibule, Selectivity Filter, and Conducting Pore.
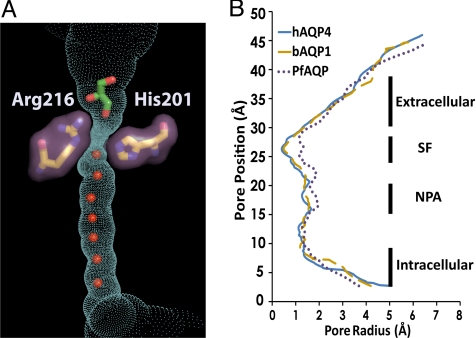
AQP4 is a water-selective channel. Signature to the water-selective channels, His-201 lies directly in the selectivity filter, reducing the channel diameter to ≈1.5 Å, sterically excluding the passage of glycerol (Fig. 2). AQP4 was purified and crystallized in the presence of 5% (vol/vol) glycerol (0.7 M), and 3 glycerol molecules are found in the extracellular vestibule, although not in the selectivity filter where the 2 glycerol-conducting AQPs, GlpF, and PfAQP, bind glycerol identically to one another (26, 27) (Fig. S5). In the water-selective rAQP1, the double mutant Phe56Ala and His180Ala (Phe-77 and His-201 in hAQP4) (Fig. S5) allows for the passage of glycerol, showing that steric occlusion is one mechanism for exclusion of larger solutes (28).
Fig. 2.
The conducting pore. The trace of the pore inner surface is shown in cyan. The selectivity filter residues, Arg-216 and His-201, are shown as sticks with surfaces in purple. The glycerol molecule is shown as green stick, and the water molecules in the channel are shown as red spheres. (B) Plot of the channel radius versus position along the pore for human AQP4, bovine AQP1 (bAQP1), and the P. falciparum AQP (PfAQP). Regions of the channel are labeled as extracellular vestibule, the selectivity filter (SF), the NPA motif, and the intracellular vestibule. The pore inner surface and its dimension are calculated using Hole 2.0 (51).
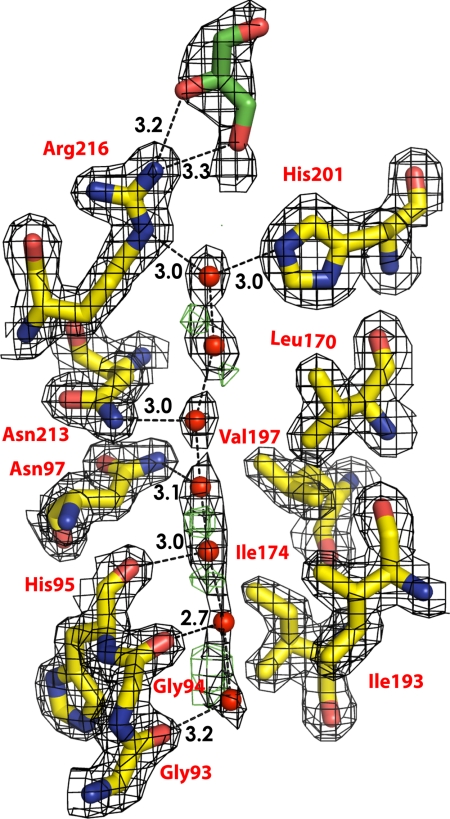
The ≈25-Å long conducting pore contains a line of water molecules and no solute molecule. However, the electron density of the water molecules are distributed along the pore with residual positive Fo − Fc density observed in between water positions indicating increased anisotropic distribution along the channel axis, implying low-energy barriers between the water molecules along the direction of the channel (Fig. 3). As in other AQPs, the pathway through the channel is amphipathic. The hydrophobic sides are formed by the side chains of Phe-77, Ile-81, Val-85, Leu-170, Ile-174, and Val-197. The 8 backbone carbonyls of Gly-93, Gly-94, His-95, and Ile-96, from the cytoplasmic side and Gly-209, Ala-210, Ser-211, and Met-212 form the hydrophilic hydrogen bond acceptors for 8 positions of water molecules in transit. This arrangement allows bidirectional conductance of water from either side of membrane.
Fig. 3.
Electron density. Residues that form the wall of the pore are shown in sticks. Water molecules are shown as red spheres. The glycerol molecule is shown in green stick. The 2Fo-Fc density is shown in black, contoured at 1.2 σ. Positive Fo − Fc density is shown in green, contoured at 3 σ. There is no negative Fo − Fc density.
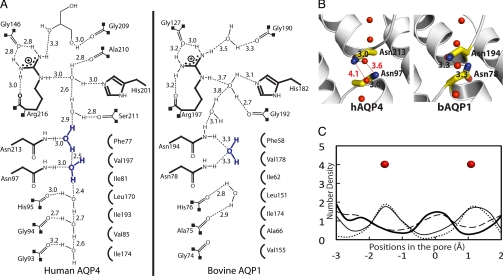
The asparagines 213 and 97 of the 2 almost totally conserved Asn-Pro-Ala (NPA) motifs form the canonical “fireman's grip-like” structure in the center of the pore (26), and provide the defining force that orients water as it passes through the midpoint of the channel. However, in hAQP4, each asparagine donates its single, highly oriented hydrogen bond to a separate water molecule (Fig. 3). This arrangement is a key variant, because in 3 other water-selective AQP structures (23, 24, 29), the 2 asparagines donate hydrogen bonds to a single water molecule.
Conductance of hAQP4 in Proteoliposomes.
Both full-length hAQP4 and trypsinized hAQP4 were reconstituted into proteoliposomes, and water and glycerol conduction were measured. The proteoliposomes containing the full-length protein conducts water at the rate of 120.3 ± 9.5 s−1, 12 times the rate for protein-free liposomes 9.80 ± 0.70 s−1, and do not conduct any glycerol, with measured rate of 0.071 ± 0.001, the same as for protein-free liposomes 0.071 ± 0.0001. Proteoliposomes containing the trypsinized protein conduct water at the rate (relative rates) of 107.4 ± 1.5 s−1 compared with 5.71 ± 0.05 s−1 for protein-free liposomes, and do not conduct glycerol with measured rates of 0.077 ± 0.002, the same as 0.071 ± 0.0001 for protein-free liposomes (Fig. S1a). Thus, both forms of AQP4 conduct water, but do not conduct glycerol. The data also demonstrate that the protein expressed in P. pastoris is functional, and that trypsinolysis of the protein does not have an apparent effect on water conductivity.
Discussion
Implication of the C Loop in Cell–Cell Adhesion and NMO.
X-ray structures of AQPs from all 3 kingdoms, bacteria, archaea, and eukaryote, have been determined. The C loop that connects M4 and M5 and forms part of the extracellular vestibule shows remarkable features across all kingdoms. GlpF is unique in that part of the C loop folds as 2 α-helices. Both AqpM (30) and the spinach AQP SoPIP2;1 have a 2-turn helix entering the C loop. PfAQP has a short helix that is very similar to its bacterial aquaglyceroporin homolog, GlpF (Fig. 1D). Among others, rAQP4 stands out in that it has a 1-turn 310 helix (Fig. 1E) and is similar to the 2-turn helix seen in AqpM and the spinach AQP SoPIP2;1 (Fig. 1F). Hiroaki et al. (13) proposed from the rAQP4 electron diffraction structure that the 310 helix in the C loop (residues 139 to 142; Fig. S4) is the main region that provides for AQP4-mediated cell–cell adhesion (13). The hAQP4 and rAQP4 share the same protein sequences in this loop (Fig. S3), but in the X-ray structure of hAQP4, even though this region has the hydrogen bonds of an α-helix, it is a well-defined loop and does not adopt the dihedral angles (ϕ, ψ) of a 310 helix (Fig. S4). Therefore, the 310 helix in the C loop is a conformation that may be induced when 2 cells expressing AQP4 on the surface are in close proximity bringing 2 C loops into direct contact. It is also possible that the 310 helix is nonphysiological, induced by the packing of the rAQP4 double-layered structure in the electron microscopy samples (Fig. S2b) (13). In 2 different cell types, each with or without expression of orthogonally arrayed AQP4, separate cell–cell adhesion and dynamic light scattering assays demonstrated that AQP4 does not have any cell–cell adhesion property (31). Because in our 1.8-Å hAQP4 structure the C loop is not involved in the crystal packing, the lack of any helix in the C loop, therefore, supports the conclusion that AQP4 does not strongly drive adhesion, and that the 310 helix in the C loop observed in the rAQP4 structure may be induced by crystal contact.
Nevertheless, given the diversity and the flexibility of the C loop, it is possible that the C loop could have an important role in the pathogenesis of NMO. It has been proposed that the 3 extracellular loops (A, C, and E) could independently or jointly form the antigenic epitopes for the NMO-IgG autoantibody (32). Therefore, it is possible that the variable conformations of the C loop could aid in the affinity of the autoantibody to AQP4, so strengthening the downstream cascades of complement-mediated inflammatory response.
Recruitment of Solutes in the Extracellular Vestibule.
In the crystal structures of GlpF and PfAQP, glycerol molecules are located in the selectivity filter, as well as in the entry vestibule (Fig. S6). In aquaglyceroporins, the vestibule may serve as a place for glycerol recruitment and desolvation of solutes for transport through the channel. For the water-specific bAQP1, the spinach AQP SoPIP2;1, bAQP0, and hAQP5, glycerol was not included in the crystallization conditions, but for the water-specific AqpZ where 5% glycerol (0.7 M) was present during crystallization, glycerol molecules were also not found in the vestibule (Fig. S6) (29). The 1.8-Å structure of the water-selective hAQP4 (crystallized in the presence of 5% glycerol), has no glycerol in the selectivity filter, but has 3 glycerol molecules nonspecifically bound in the extracellular vestibule. The extracellular vestibule, a canyon defined primarily by the A and C loop, interacts with the glycerol molecules through hydrogen bonds of the amide backbone with the hydroxyl groups of glycerol. It is tantalizing to speculate that the locations of the glycerol molecules in the vestibule could represent “fragment binding sites” for defining inhibitors that would bind from the extracellular side.
Because there are tremendous prospects for drugs that inhibit AQP4, and indeed many inhibitors of AQP4 have been described in the literature to date, we cocrystallized hAQP4 with 5 mM of 3 such compounds, tetraethylammonium (TEA), acetazolamide, and rizatriptan, and determined their cocrystal structures. In oocytes swelling assays, these compounds were reported to be AQP4 inhibitors with IC50 in the micromolar range (6–9); however, we have not been able to detect any compound bound in the structures. We then reconstituted purified hAQP4 into liposomes, and measured water conductance in the presence of these compounds. TEA up to 10 mM had no effect on hAQP4 water conductance. Acetazolamide and rizatriptan do inhibit water conductance with an approximate IC50 in the low millimolar range (≈3 and ≈1 mM, respectively; Fig. S1b). We argue that water conductance measurement using purified proteoliposomes is more reliable than in oocytes. In a similar experiment, acetazolamide was also found to inhibit rAQP4 in the mM range, but not hAQP1 (33). These results call into question the previous interpretation of TEA, acetazolamide and rizatriptan as micromolar inhibitors of AQP4.
Arginine 216 Environment Determines Conductance of Water in AQPs.
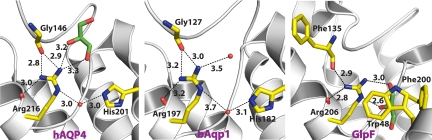
Because the glycerol conducting PfAQP conducts water as well as a water channel, whereas GlpF conducts water very poorly, we proposed that the efficiency of water conductance may be proportional to the effective neutralization of the formal positive charge on the arginine side chain by 5 hydrogen bond acceptors, 3 from the protein and 2 from the waters in transit (27). The hAQP4 is a water-specific channel, and similar to all high water conductance AQPs, the NεH, Nη1H, Nη2H of Arg-216 in the selectivity filter are all hydrogen bonded to other acceptor oxygens of the protein, leaving 2, one each from NεH and NηH as donors to the waters in transit (Fig. 4). For the water-selective bAQP1, the arginine (Arg-197) environment is very similar to that in hAQP4 (Fig. 4). However, in the glycerol conducting GlpF, with low water conductance, the selectivity filter arginine (Arg-206) has only one of the NηH satisfied (Fig. 4), so it is possible that the extra cost of desolvating the fully charged guanidinium leaves GlpF “holding on” to water molecules in transit. This higher degree of the guanidinium cation buffering by the protein may be the basis for higher efficiency water transport among the AQPs.
Fig. 4.
Comparison of the hydrogen bond network of the selectivity filter arginine of hAQP4, bAQP1, and GlpF. Protein C-alpha is shown in diagram representation. Residues of the selectivity filter and glycerol molecules are shown as sticks. Water molecules are shown as red spheres.
Variant Role of the Dual NPA Motifs in Proton Exclusion.
The interlocking of 2 almost totally conserved NPA motifs in AQPs provides 2 highly oriented donors from NδΗ2 of the 2 asparagines to the center of the channel. We proposed that they have a key role in insulation against any conduction of protons or ions while supporting a single file of hydrogen bonded water molecules throughout the entire length of the channel (34). In the crystal structures of the water-selective Escherichia coli AqpZ and bAQP1, the central water is positioned in between the 2 asparagines of the NPA motifs, and receives 2 hydrogen bonds from the 2 asparagine NδΗ2s (represented by bAQP1 in Fig. 5 A and B). Such orientation of the central water may be a factor in preventing proton conduction through the channel by the highly cooperative hop-and-turn Grotthuss relay mechanism (35, 36), because the central water cannot rotate to accept or exchange its proton (26, 37). However, the hAQP4 crystal structure does not have a water molecule centrally located between the 2 NPA motifs. Instead, each asparagine of the NPA motifs (213 and 97) donates a hydrogen bond to a different water molecule (Fig. 5). This arrangement is also observed in the crystal structures of hAQP5 and the spinach AQP SoPIP2;1. Therefore, in this subclass of AQP structures, these 2 water molecules may each be somewhat freer to rotate in a concerted fashion leaving 1 additional non-hydrogen bonded water hydrogen. These AQPs equally insulate against any proton leakage, suggesting that the 2 models for the central waters may represent intermediates that are very close in free energy.
Fig. 5.
The NPA motifs. (A) Schematic representation of the hydrogen bonding network through the channels of hAQP4 and bAQP1. The distances are between heavy-atom to heavy-atom. (B) Stick representation of the NPA motifs. Distances that are too long to be a hydrogen bond are colored in red. (C) Plot of the MD simulations of hAQP4 from 4 different experiments. Details are described in Discussion.
To see whether this observation constitutes a difference in mechanistic terms, molecular dynamic (MD) simulations were performed to assess the probability of finding a single central water molecule bonding to both NPA asparagines simultaneously. The results of the MD simulations were used to calculate the number density of water molecules along the channel near the 2 NPA motifs (Fig. 5C). Four simulations were carried out. In the first simulation (thin solid line), the system commenced from the protein and crystallographic waters; the protein positions were kept frozen throughout the simulation. The resulting number density is consistent with the observed water positions in the hAQP4 crystal structure (solid red spheres), and suggests that this particular configuration of the protein supports 2 energy minima for water in the NPA region. To test whether this result was biased by the introduction of the crystallographic water molecules into the simulation, a second simulation (dotted line) was performed, in which the protein positions were kept frozen, but the crystallographic waters were removed from the simulation, and the pore evacuated. After equilibration with the remaining simulation water molecules, the resulting number density is consistent with the experimentally observed water positions in the hAQP4 crystal structure, and gives further support that the calculation is stable and reiterates the particular observed configuration of hAQP4. In the third simulation (thick solid line), the system contained both the protein and the crystallographic waters, but instead of freezing the atomic positions of the protein, the heavy atoms (N, C, and O) were restrained to their crystallographic positions using a harmonic restraint. This slight release of the protein atoms from their crystallographic positions allows the number density to shift such that a water molecule positions itself in the middle of the NPA motifs, as is observed in the crystal structures of bAQP1 (Fig. 5 A and B; Fig. S7). Last, in the forth simulation (dashed line), all of the positional restraints were removed from the protein, allowing the protein to move freely. Again, the resulting number density suggests the presence of a water molecule in the middle of the NPA region. Thus, we conclude that the observed position of the 2 waters near the NPA region, in the hAQP4 crystal structure, do not conform to a functionally distinct variant, but rather, signal the lowered barriers to conductance between these sites.
Analogous sets of simulations were performed on bAQP1. In all 4 simulations, the number density supports the mechanism that as a water molecule travels through an AQP or aquaglyceroporin channel, it does transition into and through a central position where it simultaneously accepts hydrogen bonds from both NPA asparagine donors, and that in this transition state, the line of waters throughout the channel are polarized such that all waters have their dipoles oriented away from the central water molecule (Fig. S7).
Our first AQP structure, for GlpF, led to our suggestion that the highly ordered, polarized water orientation throughout the channel was a key factor in the exclusion of passage of protons in a fully concerted fashion without developing charge within the channel itself (34). Other considerations have also been raised. The Warshel group calculated the electrostatic barrier of placing a proton/hydronium ion into the center of the channel, and concluded that this electrostatic desolvation barrier alone excludes mass transfer of hydronium ions to the center of the channel, and that the contribution of the NPA asparagines and the helix-induced dipoles at the NPA region are an insignificant addition to a barrier for hydronium ions at this position (38–40). However, if a proton were transferred through the channel by a Grotthuss mechanism, no formal charge need ever appear within the channel because of the concerted hopping of protons, in at one side and out at the other. The Grotthuss mechanism has been assessed (41) by Poisson–Nernst–Planck theory, which indicates that there are high barriers to the Grotthuss mechanism. Thus, the cost of this degree of ordering may abrogate the Grotthuss mechanism through a line of water that is this long. Using free-energy simulations and continuum electrostatic calculations, the Roux group adds weight to the proposal that the dehydration of an ion as it approaches the channel presents a major barrier to mass transfer of an ion through the channel, but concludes that other factors including the NPA region within the channel are important as well (42). Using multistate empirical valence bond (MS-EVB) models for proton transfer, Ilan et al. (37) suggests that the bipolar line of water, the charge on the conserved arginine of the selectivity filter, and ion desolvation penalties are all relevant as we proposed initially (34).
Perspective.
Because of the flexibility of the termini, the N-terminal 19 aa and the C-terminal 64 aa were removed from the M1 isoform by trypsinolysis, as determined by MS and N-terminal sequencing. Thus, the crystal structure we report at 1.8-Å resolution contained Glu-20 to Lys-259 with visible electron density from Gln-32 to Pro-254. In contrast, the electron diffraction structure of rAQP4 was expressed as the M23 isoform, and revealed density for residues 31–254, consistent with a natively unstructured region at each of the termini. Thus, whereas the structure fully elucidates all features of the water transport pathway, it does not yet address the question of how the M1 isoform works together with the M23 isoform to limit the extent of orthogonal array formation. It also remains to be determined how the C terminus interacts with α-syntrophin in the context of polarized expression in astrocytes.
Questions also remain on the mechanism of how Ser-111 and Ser-180 phosphorylation affect AQP4 conductance. Studies have shown that the phosphorylation state of Ser-111 on the B loop and Ser-180 on the D loop affect water conductance of AQP4 (Fig. S3) (18–20). Phosphorylated Ser-180 has been speculated to interact with Lys-259, Arg-260, and Arg-261 as a means of tethering the C-terminal domain to block the channel (13), and phosphorylated Ser-111 has been proposed to mimic the gating mechanism of the spinach AQP SoPIP2;1 by disrupting the network of hydrogen bonds that anchor the D loop to the N terminus: thus, opening the channel (20). In our 1.8-Å crystal structure, the channel is open and, although the density for the B and D loops is clear, no phosphate is observed on either serine. Because the protein was trypsinized, neither the N- and C-terminal domains are present. Although we cannot comment on the interaction of Ser-180 with the C-terminal domain, we think it is unlikely that the gating of AQP4 at Ser-111 is similar to SoPIP2;1, because the D loop of hAQP4 is 4 residues shorter than in SoPIP2;1, and residues in the N-terminal domain that are involved in binding to the D loop (Asp-28 and Glu-31) are different amino acids in hAQP4 (Fig. S3).
The high resolution structure of the phosphorylated protein, the M1 isoform, and complexes with binding elements of syntrophin may address these issues of larger scale assemblies beyond that of the tetrameric AQP4.
Materials and Methods
Materials and methods for protein expression and purification, crystallization, proteoliposome assay, and MD simulations are described in SI Materials and Methods.
For data collection and model building, diffraction data were collected using a wavelength of 1.11 Å at Beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory). Data were processed by using HKL2000 (43). Molecular replacement was performed with Phaser (44) using the rAQP4 electron diffraction structure (PDB code: 2D57) (13), as a search model. Subsequent iterative cycles of manual building and retrained refinement were done using Coot (45) and Refmac5 in CCP4 (46). TLS refinement in Refmac5 was applied in the last stage using 19 TLS groups obtained from the TLS analysis server (Table S1 and Table S2) (47, 48). Riding hydrogen atoms were generated during refinement, but not written to the output. Structure was assessed using PROCHECK (49) and MolProbity (50). Data processing and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P4212 |
| Cell dimensions a, c, Å | 82.1, 76.4 |
| Resolution, Å | 1.8 (1.86–1.8) |
| Rsym | 9.0 (79.6) |
| I/σI | 20 (3.3) |
| Completeness, % | 99.9 (100) |
| Redundancy | 11.9 (12.0) |
| Refinement | |
| Resolution, Å | 36–1.8 |
| No. of unique reflections | 23,583 |
| Rwork/Rfree | 16.0/16.5 |
| No. of atoms | |
| Protein | 1,667 |
| Glycerol | 30 |
| β-OG | 20 |
| Water | 61 |
| Avg B-factors, Å2 | |
| Protein | 27 |
| Glycerol | 42 |
| β-OG | 55 |
| Water | 45 |
| rmsd | |
| Bond lengths, Å | 0.01 |
| Bond angles, ° | 1.047 |
Values in parentheses are for the highest resolution shell. Rfree is calculated from 5% of reflections chosen randomly.
Supplementary Material
Acknowledgments.
We thank Joseph Luft and Jennifer Wolfley (Hauptman-Woodward Medical Research Institute) for setting up the drop-volume ratio crystallization trials of the full-length AQP4 and Liang Li (Ismagilov Laboratory, University of Chicago) for his help in the capillary microbatch crystallization trials of the same (both produced crystals although neither yielded better than the 8-Å resolution we obtained for the full-length molecule); and John Lee and Zach Newby for helpful comments on the manuscript. We thank James Holton for his assistance at the Advanced Light Source Beamline 8.3.1, which was supported by National Institutes of Health (NIH) Grant GM074929 (to R.M.S.). This work was supported by NIH Grant GM24485 (to R.M.S.) and by NIH Roadmap Center Grant P50 GM073210. I.C. was supported from a scientific computing grant from Microsoft Corporation (to R.M.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 3GD8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902725106/DCSupplemental.
References
- 1.Rojek A, Praetorius J, Frokiaer J, Nielsen S, Fenton RA. A current view of the mammalian aquaglyceroporins. Annu Rev Physiol. 2008;70:301–327. doi: 10.1146/annurev.physiol.70.113006.100452. [DOI] [PubMed] [Google Scholar]
- 2.King LS, Kozono D, Agre P. From structure to disease: The evolving tale of aquaporin biology. Nat Rev Mol Cell Biol. 2004;5:687–698. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 3.Badaut J, Brunet JF, Regli L. Aquaporins in the brain: From aqueduct to “multi-duct.”. Metab Brain Dis. 2007;22:251–263. doi: 10.1007/s11011-007-9057-2. [DOI] [PubMed] [Google Scholar]
- 4.Manley GT, et al. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat Med. 2000;6:159–163. doi: 10.1038/72256. [DOI] [PubMed] [Google Scholar]
- 5.Badaut J, Lasbennes F, Magistretti PJ, Regli L. Aquaporins in Brain: Distribution, Physiology, and Pathophysiology. J Cereb Blood Flow Metab. 2002;22:367–378. doi: 10.1097/00004647-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Detmers FJM, et al. Quaternary ammonium compounds as water channel blockers: Specificity, potency, and site of action. J Biol Chem. 2006;281:14207–14214. doi: 10.1074/jbc.M513072200. [DOI] [PubMed] [Google Scholar]
- 7.Huber VJ, Tsujita M, Yamazaki M, Sakimura K, Nakada T. Identification of arylsulfonamides as Aquaporin 4 inhibitors. Bioorg Med Chem Lett. 2007;17:1270–1273. doi: 10.1016/j.bmcl.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Huber VJ, Tsujita M, Kwee IL, Nakada T. Inhibition of Aquaporin 4 by antiepileptic drugs. Bioorg Med Chem. 2009;17:418–424. doi: 10.1016/j.bmc.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 9.Huber VJ, Tsujita M, Nakada T. Identification of aquaporin 4 inhibitors using in vitro and in silico methods. Bioorg Med Chem. 2009;17:411–417. doi: 10.1016/j.bmc.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 10.Yang B, Zhang H, Verkman AS. Lack of aquaporin-4 water transport inhibition by antiepileptics and arylsulfonamides. Bioorg Med Chem. 2008;16:7489–7493. doi: 10.1016/j.bmc.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingerchuk DM, Lennon VA, Lucchinetti CF, Pittock SJ, Weinshenker BG. The spectrum of neuromyelitis optica. Lancet Neurol. 2007;6:805–815. doi: 10.1016/S1474-4422(07)70216-8. [DOI] [PubMed] [Google Scholar]
- 12.Furman CS, et al. Aquaporin-4 square array assembly: Opposing actions of M1 and M23 isoforms. Proc Natl Acad Sci USA. 2003;100:13609–13614. doi: 10.1073/pnas.2235843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiroaki Y, et al. Implications of the aquaporin-4 structure on array formation and cell adhesion. J Mol Biol. 2006;355:628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- 14.Michele DE, Campbell KP. Dystrophin-glycoprotein complex: Post-translational processing and dystroglycan function. J Biol Chem. 2003;278:15457–15460. doi: 10.1074/jbc.R200031200. [DOI] [PubMed] [Google Scholar]
- 15.Amiry-Moghaddam M, Frydenlund DS, Ottersen OP. Anchoring of aquaporin-4 in brain: Molecular mechanisms and implications for the physiology and pathophysiology of water transport. Neuroscience. 2004;129:997–1008. doi: 10.1016/j.neuroscience.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa H, Ma T, Skach W, Matthay MA, Verkman AS. Molecular cloning of a mercurial-insensitive water channel expressed in selected water-transporting tissues. J Biol Chem. 1994;269:5497–5500. [PubMed] [Google Scholar]
- 17.Savage DF, Stroud RM. Structural basis of aquaporin inhibition by mercury. J Mol Biol. 2007;368:607–617. doi: 10.1016/j.jmb.2007.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Z, Wax MB, Patil RV. Regulation of aquaporin-4 water channels by phorbol ester-dependent protein phosphorylation. J Biol Chem. 1998;273:6001–6004. doi: 10.1074/jbc.273.11.6001. [DOI] [PubMed] [Google Scholar]
- 19.Zelenina M, Zelenin S, Bondar AA, Brismar H, Aperia A. Water permeability of aquaporin-4 is decreased by protein kinase C and dopamine. Am J Physiol Renal Physiol. 2002;283:F309–318. doi: 10.1152/ajprenal.00260.2001. [DOI] [PubMed] [Google Scholar]
- 20.Gunnarson E, et al. Identification of a molecular target for glutamate regulation of astrocyte water permeability. Glia. 2008;56:587–596. doi: 10.1002/glia.20627. [DOI] [PubMed] [Google Scholar]
- 21.Hedfalk K, et al. Aquaporin gating. Curr Opin Struct Biol. 2006;16:447–456. doi: 10.1016/j.sbi.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Tornroth-Horsefield S, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 23.Sui H, Han B-G, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 24.Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci USA. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsefield R, et al. High-resolution x-ray structure of human aquaporin 5. Proc Natl Acad Sci USA. 2008;105:13327–13332. doi: 10.1073/pnas.0801466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D, et al. Structure of a Glycerol-Conducting Channel and the Basis for Its Selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 27.Newby ZER, et al. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat Struct Mol Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc Natl Acad Sci USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savage DF, Egea PF, Robles-Colmenares Y, O'Connell JD, III, Stroud RM. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JK, et al. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc Natl Acad Sci USA. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Verkman AS. Evidence against involvement of aquaporin-4 in cell-cell adhesion. J Mol Biol. 2008;382:1136–1143. doi: 10.1016/j.jmb.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graber DJ, Levy M, Kerr D, Wade WF. Neuromyelitis optica pathogenesis and aquaporin 4. J Neuroinflammation. 2008;5:22. doi: 10.1186/1742-2094-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanimura Y, Hiroaki Y, Fujiyoshi Y. Acetazolamide reversibly inhibits water conduction by aquaporin-4. J Struct Biol. 2009;166:16–21. doi: 10.1016/j.jsb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Tajkhorshid E, et al. Control of the Selectivity of the Aquaporin Water Channel Family by Global Orientational Tuning. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 35.Grotthuss CJTd. Memoir on the decomposition of water and of the bodies that it holds in solution by means of galvanic electricity (Translated from French) Ann Chim. 1806;58:54–74. [Google Scholar]
- 36.Grotthuss CJTd. Memoir on the decomposition of water and of the bodies that it holds in solution by means of galvanic electricity. 1805. Biochim Biophys Acta. 2006;1757:871–875. doi: 10.1016/j.bbabio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Ilan B, Tajkhorshid E, Schulten K, Voth GA. The mechanism of proton exclusion in aquaporin channels. Proteins. 2004;55:223–228. doi: 10.1002/prot.20038. [DOI] [PubMed] [Google Scholar]
- 38.Kato M, Pisliakov AV, Warshel A. The barrier for proton transport in aquaporins as a challenge for electrostatic models: The role of protein relaxation in mutational calculations. Proteins. 2006;64:829–844. doi: 10.1002/prot.21012. [DOI] [PubMed] [Google Scholar]
- 39.Burykin A, Warshel A. What really prevents proton transport through aquaporin? Charge self-energy versus proton wire proposals. Biophys J. 2003;85:3696–3706. doi: 10.1016/S0006-3495(03)74786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burykin A, Warshel A. On the origin of the electrostatic barrier for proton transport in aquaporin. FEBS Lett. 2004;570:41–46. doi: 10.1016/j.febslet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, et al. Charge delocalization in proton channels, I: The aquaporin channels and proton blockage. Biophys J. 2007;92:46–60. doi: 10.1529/biophysj.106.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti N, Tajkhorshid E, Roux B, Pomès R. Molecular basis of proton blockage in aquaporins. Structure. 2004;12:65–74. doi: 10.1016/j.str.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.CCP4 Collaborative Computational Project N. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 48.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 50.Lovell SC, et al. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 51.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.