Figure 2. Transmembrane 5 segment mutations enhance GlpG protease activity in E. coli cells.
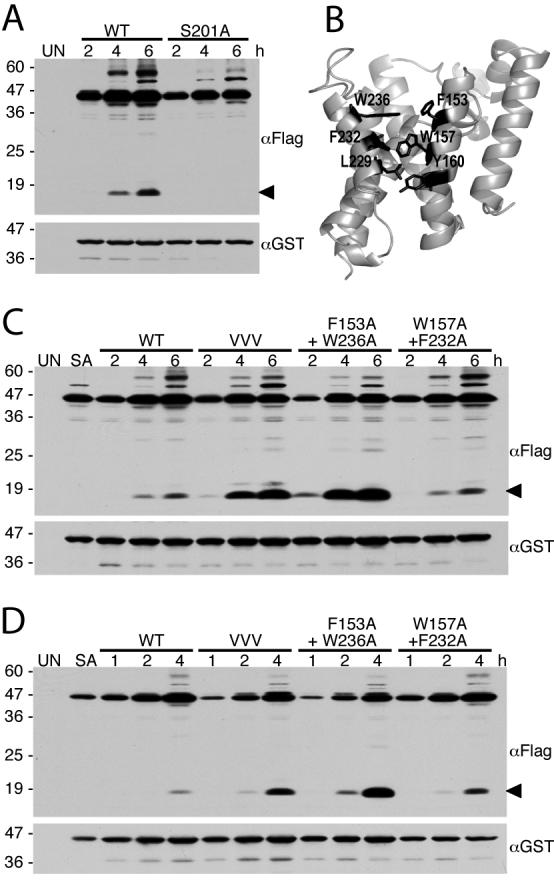
A. Western analysis of Spitz substrate processing (anti-Flag) and GlpG protease expression (anti-GST) in E. coli cells following 2, 4 and 6 hours post-induction. Uninduced E. coli cells (UN) were used as a negative control. The black arrow on the right denotes the cleavage product. Higher molecular weight substrate bands likely represent aggregates. Protein mass standards (in kDa) are depicted to the left of the images. B. Back view of GlpG (open conformation, PDB N2RF) with transmembrane residues on helix 2 and 5 highlighted in black. C and D. Western analysis of cultures expressing wildtype or GlpG helix 5 mutant enzymes with the Spitz substrate revealed a dramatic increase of protease activity of the mutant GlpG enzymes. VVV is the triple transmembrane helix 5 mutant L229V+F232V+W236V. Note that wildtype and mutant-expressing cultures were grown and analyzed in parallel, with data in C and D being from two independent experiments (samples in D were analyzed at shorter times post-induction). UN and SA denote uninduced wildtype GlpG and S201A GlpG mutant cultures, respectively, that served as general negative controls.
