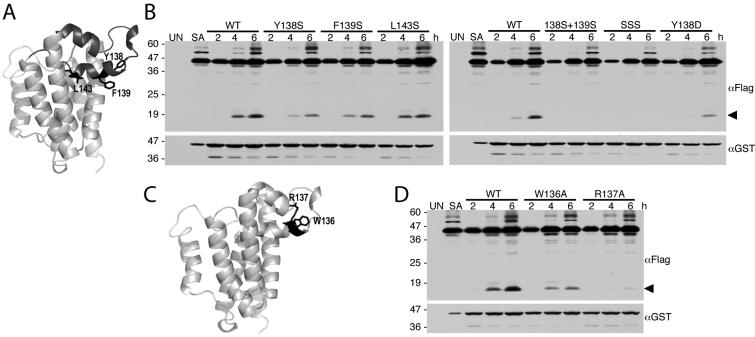
Figure 5. Mutations within the L1 loop decrease GlpG protease activity in E. coli cells.
A.Lateral view of GlpG, with the membrane-inserted L1 loop highlighted in black. Sidechains of residues Y138, F139, and L143 that would be expected to contact membrane lipid and thus stabilize the membrane inserted conformation of the loop are shown. B. Western analysis of cultures expressing wildtype and L1 loop mutant GlpG enzymes with the Spitz substrate. Anti-flag was used to reveal the Spitz substrate while GlpG protease expression levels were monitored using anti-GST. The black arrow denotes the cleavage product. Note that all L1 loop mutants decrease GlpG protease activity. C. Lateral view of the GlpG structure (PDB N2RF) highlighting the W136 and R137 residue pair (in black) within the L1 loop. D. Western analysis of cultures co-expressing wildtype versus W136A or R137A mutant GlpG with the Spitz substrate. Note that the R137A mutation strongly decreased GlpG protease activity. In all B and D panels, UN and SA denote uninduced wildtype and S201A-expressing cultures, and location of protein mass standards (in kDa) are depicted on the left.