Figure 6. Mutation of non-catalytic residues lining the GlpG active site hinder protease activity.
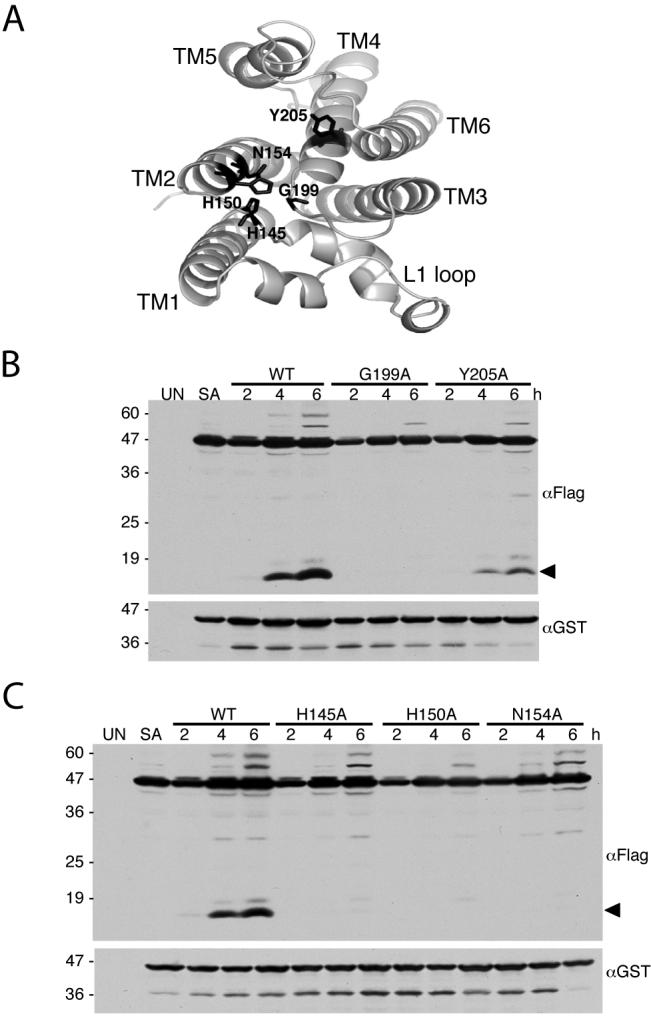
A. Top view of GlpG (PDB N2RF), with the L1 loop down. Side-chains of conserved non-catalytic active sites residues under investigation are highlighted in black. B and C. Western analysis of cultures expressing either wildtype of mutant GlpG proteases with the Spitz substrate. All mutant enzymes show a decrease in substrate processing (cleavage product denoted by black arrow). UN and SA denote uninduced wildtype and S201A-expressing cultures, respectively. Location of protein mass standards (in kDa) are depicted on the left.
