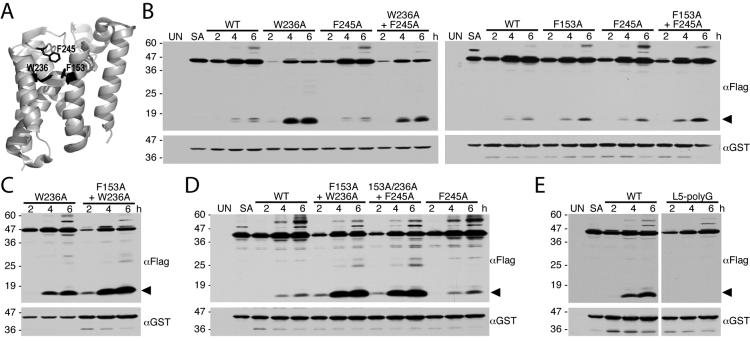
Figure 7. Single and combined Helix 2, Helix 5 and Cap mutations enhance GlpG protease activity in E. coli cells.
A. Lateral back view of the closed conformation of GlpG (PDB 2IC8) showing the sidechain (in black) interactions of residues W236 on helix 5, F153 on helix 2, and F245 on the L5 Cap. B. Western analysis of bacteria expressing wildtype, single and double mutants of GlpG and the Spitz substrate. Note that, of the single mutants, the most profound increase in protease activity was observed with the transmembrane helix 5 mutant W236A. C. Combining the F153A mutant with the W236A mutant increases GlpG protease activity relative to the single W236A mutant in E. coli cells. D. Combining the Cap mutant F245A with the double transmembrane helix mutant F153A+W236A did not increase the GlpG protease activity of the triple mutant relative to that of the F153A+W236A double mutant in bacterial cells. E. Removing the sidechains of Cap residues 243-250 by mutating all to glycine (L5-polyG) abolished GlpG protease activity in E. coli cells. In all panels, the cleavage product is denoted by a black arrow, UN and SA are uninduced wildtype and S201A-expressing cultures, respectively, and the location of protein mass standards (in kDa) are depicted on the left.