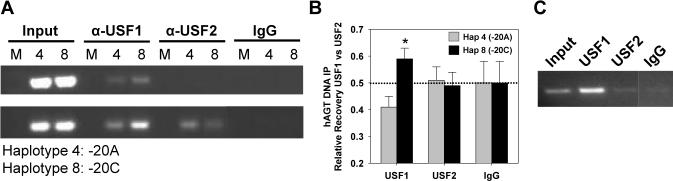
Figure 3. Plasmid and Chromatin Immunoprecipitation.
A. HepG2 cells were mock transfected (first in each set of three lanes) or transfected with AGT reporter vectors containing −20A (haplotype 4) or −20C (haplotype 8). IP was carried out using the indicated antibodies followed by PCR using primers specific for the plasmid DNA. Results from two independent experiments are shown. B. Summary of HepG2 PIP experiments quantified by a real-time PCR SNP assay. The relative recovery of −20A (gray) and −20C (filled) DNA IP by the indicated antisera is shown. ΔCT values were corrected for input, converted to a fraction of total (USF1/USF1+USF2 or USF2/USF1+USF2), and are presented as mean±SEM. *, P<0.05 for −20C vs −20A (n=5). The dotted line represents equal recovery of −20A vs −20C. C. ChIP using input, USF1, USF2 or IgG antibody control from CCF cell chromatin amplified with primers specific for hAGT. The IgG was cut from the same gel as the other lanes.