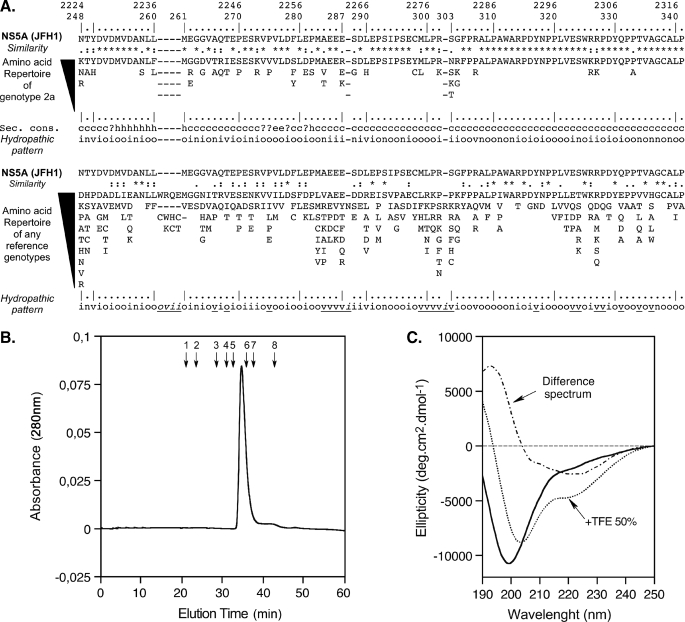
FIGURE 1.
Sequence analysis and biochemical characterization of NS5A-D2 domain 2 from HCV. A, amino acid repertoires. The NS5A-D2 aa 248–341 sequence from the HCV JFH1 strain of genotype 2a (GenBank™ accession number AB047639), which was used in this study, is indicated. Amino acids are numbered with respect to NS5A and the HCV JFH1 polyprotein (top row). The hyphens indicate the aa deletions compared with the sequence alignment of any genotypes (see below). The aa repertoire deduced from the ClustalW multiple alignments of 21 NS5A sequences of genotype 2a is shown at the top. Amino acids observed at a given position less than twice were not included. At the bottom is the aa repertoire of the 27 representative NS5A sequences from confirmed HCV genotypes and subtypes (listed with accession numbers in Table 1 in Ref. 71; see the European HCV Database for details). The degree of aa and physicochemical conservation at each position can be inferred from the extent of variability (with the observed aa listed in decreasing order of frequency from top to bottom) together with the similarity index according to ClustalW convention (asterisk, invariant; colon, highly similar; dot, similar (40)) and the consensus hydropathic pattern deduced from the consensus aa repertoire: o, hydrophobic position (Phe, Ile, Trp, Tyr, Leu, Val, Met, Pro, Cys); n, neutral position (Gly, Ala, Thr, Ser); i, hydrophilic position (Lys, Gln, Asn, His, Glu, Asp, Arg); v, variable position (i.e. when both hydrophobic and hydrophilic residues are observed at a given position). A position that is underlined in the hydropathic pattern for any reference genotypes (bottom) indicates the change of position status when compared with the hydropathic pattern for genotype 2a (middle). Secondary structure predictions of NS5A-D2 are indicated as helical (h), extended (e), undetermined (coil (c)), or ambiguous (?). Sec. Cons., consensus of protein secondary structures predictions for NS5A-D2 from JFH1 strain deduced from a large set of prediction methods available at the NPSA website, including DSC, HNNC, MLRC, PHD, Predator, SOPM, and SIMPA96 available at the NPSA website (see Ref. 38 and references therein). B, gel filtration analysis of NS5A-D2 was performed on a Superdex S200 column equilibrated in 50 mm sodium phosphate, pH 7.4, 1 mm Tris(2-carboxyethyl) phosphine hydrochloride with a flow rate of 0.5 ml/min. Elution volumes of globular protein standards are indicated by black arrows with the following corresponding molecular masses: 1, thyroglobulin (669,000 Da); 2, ferritin (44,0000 Da); 3, aldolase (158,000 Da); 4, conalbumin (75,000 Da); 5, ovalbumin (43,000 Da); 6, chymotrypsin (25,000 Da); 7, ribonuclease (13,700 Da); 8, vitamin B12 (1,355 Da). C, Far-UV circular dichroism analysis of 8 μm NS5A-D2 in 10 mm sodium phosphate, pH 7.4, 1 mm Tris(2-carboxyethyl) phosphine hydrochloride (solid line) complemented with 50% TFE (dotted line). The difference spectrum (alternating dashed line) was obtained by subtracting the latter spectrum from the former.