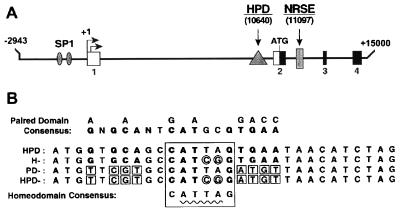
Figure 1.
(A) Location of DNA control elements in the mouse L1 gene. The first four exons are indicated by boxes labeled 1–4. The transcription initiation sites are demarcated by rightward pointing arrows. Upstream of this position is the L1 promoter containing two SP1 motifs. An NRSE that binds to the factor REST/NRSF is located within the second intron, and the HPD is located in the first intron. (B) Sequences of HPD, H−, PD−, and HPD− probes. Drawn above the HPD is the consensus paired domain binding sequence. Nucleotide similarities between this consensus sequence and the HPD are indicated in boldface type. Mutations in the PD− probe are indicated by small squares. Drawn below the PD− probe is the consensus homeodomain binding sequence CATTAG. The location of this motif in all four probes motif is indicated by a large box. Mutations made within the ATTA core sequence in the HD− variant are indicated by small circles. Both sets of mutations were included in the HPD− probe.