Figure 1.
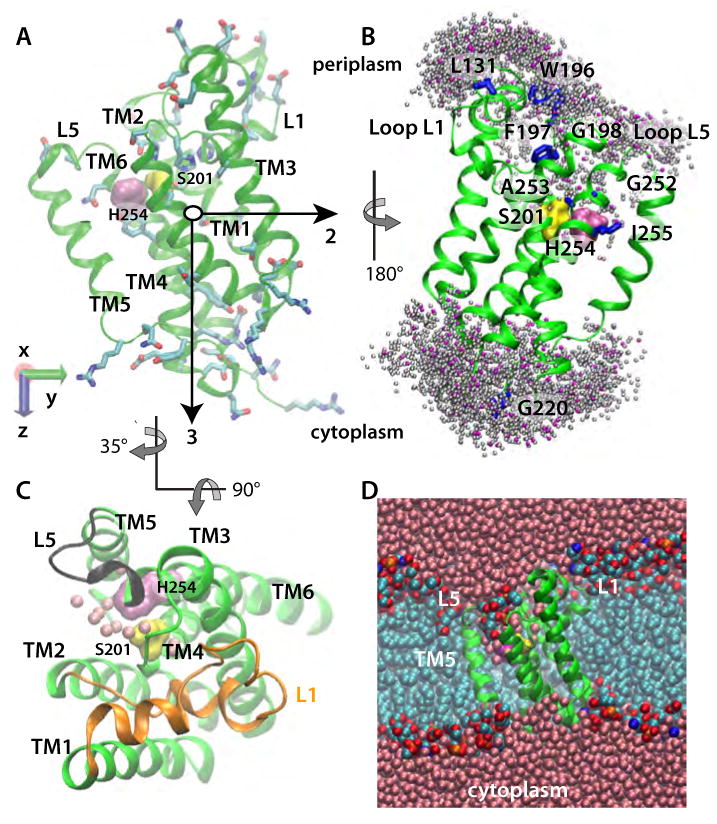
E. Coli GlpG in a POPE lipid bilayer (Sim2). (A) Starting geometry and principal axes (black arrows) of GlpG. S201 and H254 are shown as yellow and mauve surfaces, respectively; amino acid sidechains with H-bonding capability are depicted as bonds. (B) Dynamic water distribution in the active site. Locations of the water-molecule oxygen atoms located within 5 Å of the protein in a snapshot from Sim2 are shown as pink spheres. Shown in grey are water locations sampled during ten equally spaced snapshots during a subsequent 1ns segment of Sim2. Amino acids known from experiments to be accessible to bulk water (Maegawa et al., 2007) are shown as blue bonds, or as surfaces (S201 and H254). (C) View from the luminal side of GlpG from Sim2, depicting water molecules within 5Å of S201 and H254 as pink spheres, L1 in orange, and L5 in black. (D) Cut away view of GlpG in a POPE lipid bilayer, with the protein shown in green, bulk-water oxygen atoms in pink, lipid alkyl carbon cyan, lipid phosphorus orange, lipid oxygen red, and lipid nitrogen atoms blue.
