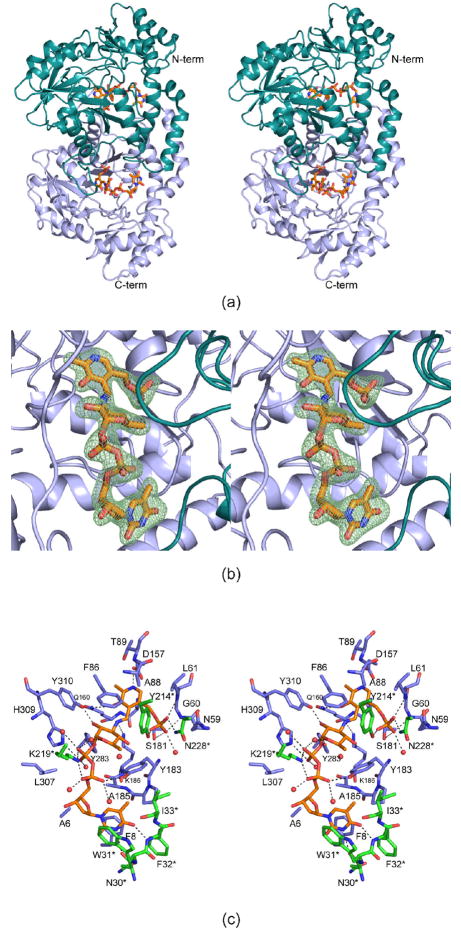
Figure 1.
Molecular architecture of QdtB. A ribbon representation of the QdtB dimer is displayed in (a). Subunits 1 and 2 of the dimer are highlighted in light blue and cyan, respectively. The ligands are drawn in stick representations. Electron density corresponding to the Schiff base between the PLP cofactor and the dTDP-sugar is presented in (b). The map was calculated with coefficients of the form (Fo−Fc), where Fo was the native structure factor amplitude, and Fc was the calculated structure factor amplitude. Atoms corresponding to the PLP and the dTDP-sugar were omitted from the calculation. A close-up view of the active site is shown in (c). Only those residues located within ~3.8 Å of the PLP and the dTDP-sugar are depicted. Amino acid residues corresponding to subunits 1 and 2 are colored in blue and green, respectively whereas the PLP and the dTDP-sugar are shown in gold bonds. Water molecules are represented as red spheres. Possible hydrogen bonding interactions are indicated by the dashed lines. All figures were prepared with the software package PyMOL (25).