Summary
Fertilization is accompanied by the construction of an extracellular matrix that protects the new zygote. In sea urchins, this structure is built from glycoproteins residing at the egg surface and in secretory vesicles at the egg cortex. Four enzymatic activities are required for the transformation of these proteins into the mechanically and chemically resilient fertilization envelope: proteolysis, transamidation, NADPH-dependent oxidation and peroxidation. Here, we identify the Strongylocentrotus purpuratus enzymes responsible for the formation of ε(γ-glutamyl)lysine crosslinks (transamidation). We find that these two transglutaminases are activated by local acidification and act on specific substrates within the fertilization envelope (including ovoperoxidase, rendezvin and SFE9). Surprisingly, these enzymes also regulate dityrosine crosslinking both by direct conjugation of ovoperoxidase and by modulating hydrogen peroxide production. Together, these results emphasize how transglutaminases can coordinate the activities of other enzymes during extracellular matrix transmogrifications.
Keywords: Extracellular matrix, Fertilization, Transamidation, Transglutaminase, Sea urchin
INTRODUCTION
Transamidation occurs in all organisms during development and in adult homeostasis (Griffin et al., 2002; Lorand and Graham, 2003), with individual transglutaminases (TGs) acting on both intra- and extracellular substrates (Aeschlimann and Thomazy, 2000). These enzymes are involved in G-protein-mediated cell signaling (Mhaouty-Kodja, 2004), keratinization of cornified epithlia (Candi et al., 2005; Lorand and Graham, 2003), insolubilization of the cell wall in the alga Chlamydomonas reinhardtii (Waffenschmidt et al., 1999), hardening of the chitinous cuticle of arthropods (Iijima et al., 2005) and the stabilization of proteins required for clotting (Chen et al., 2005; Karlsson et al., 2004; Lorand and Graham, 2003; Piacentini et al., 2000; Verderio et al., 2004). During mammalian blood clotting, for example, the transglutaminase factor XIII crosslinks serum fibrin to other extracellular matrix proteins at injury sites (Esposito and Caputo, 2005). Factor XIII proenzymes are stored as homodimers of A subunits (XIIIa) in platelets or as circulating heterotetramers of 2A and 2B subunits (XIIIa:XIIIb) in serum (Adany and Bardos, 2003). Platelet activation releases thrombin, the protease that unleashes factor XIII activity by exposing the catalytic site of this transglutaminase, whereas free Ca2+ regulates catalysis by controlling substrate access to its active cysteine (Adany and Bardos, 2003; Esposito and Caputo, 2005).
Transglutaminases are also functional at fertilization, participating in egg extracellular matrix modification. ε(γ-glutamyl)lysine crosslinks are the major contributor to teleost chorion hardening (Chang et al., 2002; Ha and Iuchi, 1998; Oppen-Berntsen et al., 1990; Yamagami et al., 1992), and are associated with morphological changes on the surface of the sea urchin fertilization envelope (Battaglia and Shapiro, 1988; Chandler and Kazilek, 1986; Larabell and Chandler, 1991; Mozingo and Chandler, 1991; Veron et al., 1977). The fertilization envelope establishes a physical block to polyspermy that protects the zygote from environmental and microbial agents (Kay and Shapiro, 1985; Wong and Wessel, 2006a). This extracellular matrix is constructed from two populations of glycoproteins, the nascent egg extracellular matrix (the vitelline layer) and the contents of cortical secretory vesicles (cortical granules) hemifused to the egg plasma membrane (Wong et al., 2007).
Four major enzymatic activities are essential for the proper assembly of the fertilization envelope: proteolysis, transamidation, hydrogen peroxide synthesis and peroxidase-dependent dityrosine crosslinking. The serine protease CGSP1 (cortical granule serine protease 1), which is derived from the cortical granules, cleaves linkages between the vitelline layer and the plasma membrane, thereby releasing the fertilization envelope from the cell surface (Carroll and Epel, 1975; Haley and Wessel, 1999). CGSP1 also regulates the activity of another cortical granule enzyme, ovoperoxidase (Haley and Wessel, 2004), which is responsible for establishing dityrosine crosslinks between adjacent proteins (Foerder and Shapiro, 1977; Kay and Shapiro, 1987; LaFleur et al., 1998; Wong and Wessel, 2008). Hydrogen peroxide, an essential substrate for ovoperoxidase activity, is synthesized from free oxygen by the NADPH reductase domain of the dual oxidase Udx1 (urchin dual oxidase 1) (Foerder et al., 1978; Warburg, 1908; Wong et al., 2004). Together, Udx1 and ovoperoxidase crosslink the proteins found throughout the fertilization envelope, thereby establishing the permeability of this zygotic extracellular matrix (Kay and Shapiro, 1985; Veron et al., 1977; Wong and Wessel, 2008). Inter-protein crosslinking occurs in two discrete subdomains of the fertilization envelope: first at the microvillar casts, the segments of the vitelline layer that cover the egg microvilli, followed by the intercast region, the area between the microvillar casts (Chandler and Kazilek, 1986; Larabell and Chandler, 1991; Mozingo and Chandler, 1991). Transamidation of proteins localized at microvillar casts is associated with the morphological change from a rounded (`igloo') to a pointed (`tent') shape (e.g. the I-to-T transition), and for fertilization envelope thickening (Battaglia and Shapiro, 1988; Chandler and Kazilek, 1986; Larabell and Chandler, 1991; Mozingo and Chandler, 1991; Veron et al., 1977), whereas the intercast region is predominantly crosslinked by ovoperoxidase (Larabell and Chandler, 1991; Mozingo and Chandler, 1991). Unlike the other enzyme activities, the transglutaminases responsible for these morphological changes remains undefined.
MATERIALS AND METHODS
Animals
Strongylocentrotus purpuratus gametes were obtained and handled as previously described (Wong and Wessel, 2004).
RNA in situ hybridization
Antisense digoxigenin-labeled RNA probes (DIG RNA Labeling Kit (SP6/T7); Roche Diagnostics Corporation, Indianapolis, IN, USA) representing S. purpuratus eTG (nucleotides 1274 to 1802) and nTG (nucleotides -62 to 403) or negative control probe (neomycin phosphotransferase II) were used at 0.1 μg probe/ml to detect native transcript in oocytes according to previously published protocols (Arenas-Mena et al., 2000).
Quantitative PCR
Real-time quantitative PCR was conducted on about 0.1 μg-equivalents of total RNA isolated from hand-collected oocytes or eggs (Song et al., 2006). Primers representing the 3′-end of each transglutaminase open reading frame were used to amplify products for eTG (199 bp using F=5′ CACTGAGGATTTGATGGTTGG with R=5′ GGTTGCCTGTCTTCTTTGGT) and nTG (169 bp using F=5′ CCACCGGAGATGAAGACTGA with R=5′ TTGCCCTTTGAAGGAACATC), and cycle numbers were normalized to ubiquitin (∼147 bp using F=5′ CACAGGCAAGACCATCACAC; R=5′ GAGAGAGTGCGACCATCCTC). Each PCR reaction was run off of cDNA (TaqMan Reverse Transcription kit; Applied Biosystems, Foster City, CA, USA) using Platinum SBYR Green qPCR SuperMix-UDG with ROX (Invitrogen Corporation, Carlsbad, CA, USA) on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
Production of polyclonal antisera
Antiserum was raised in rabbits against bacterially overexpressed, recombinant fragments subcloned downstream of a 6×-Histidine tag found in the custom-made expression vector pHisV5C4 modified from the pTAT vector (Nagahara et al., 1998). Independent antisera were raised against each transglutaminase as described previously (Wong and Wessel, 2004). Anti-eTG was generated against amino acid residues 1 to 195 (MVRR...ILFS), whereas anti-nTG was generated against residues 1 to 751 (META... IEIP), with the predicted initiation codon (methionine, ATG) designated as residue 1.
Affinity purification was accomplished against respective recombinant immunogen (residues 1-195 of eTG and 1-188 of nTG) immobilized using the Pierce AminoLink Plus Immobilization Kit (ThermoFisher Scientific Incorporated, Rockford, IL, USA) under denaturing conditions [alkaline coupling buffer with 4 M urea (pH 10)] according to manufacturer instructions. Total IgGs were isolated from heat-inactivated preimmune or anti-serum using Protein A-sepharose (Sigma-Aldrich, St Louis, MO, USA), and eluted with acidic 100 mM glycine (pH 2.5) into fourfold excess 1 M Tris (pH 8). Antiserum IgGs were then passed over the respective immunogen-immobilized resin. Bound IgGs were eluted as above, and then buffer exchanged to sterile 50 mM KCl (pH 8.0) using Amicon Ultra filters (Millipore Corporation, Billerica, MA, USA). Each IgG population was stored at -20°C as a 2 mg/ml stock in 50% glycerol.
Biochemical fractionation of eggs
Eggs were dejellied in ASW acidified with HCl (pH 5.2) followed by extensive re-equilibration to ASW (pH 8.0). These eggs were then used to isolate various fractions: (1) whole cell lysate was obtained by homogenizing cells in SDS-PAGE sample buffer; (2) cell surface complex was isolated using previously published methods (Detering et al., 1977; Kinsey, 1986), with the addition of Complete Mini EDTA-free Protease Inhibitor Cocktail (1 tablet per 10 ml ASW; Roche Diagnostics Corporation, Indianapolis, IN, USA) to the buffer. The complex was then solubilized in 1× SDS-sample buffer; (3) biochemical fractions were obtained sequentially. First, eggs were incubated for 15 minutes in 10 mM dithiothreitol (DTT; Sigma-Aldrich, St Louis, MO, USA) dissolved in ASW neutralized to pH 8.0 with Tris-base. The ASW was collected and saved as `DTT-released' protein, representing up to one-half of the chemically sensitive egg surface proteins (Carroll et al., 1977; Epel et al., 1970). The remaining cells were washed three times in ASW, and then submerged into an equal volume of 20 μg/ml A23187 in ASW (Sigma-Aldrich, St Louis, MO, USA) for 10 minutes to trigger CG exocytosis. Again, the ASW was collected and saved, representing the `exudate', which comprises the remaining egg surface proteins plus all the contents of the CGs (Carroll et al., 1977). The DTT-released and exudate samples were dialyzed extensively against TE [1 mM Tris, 1 mM EDTA (pH 8.0)], then lyophilized and resuspended in distilled water; (4) zygotic seawater was collected 20 minutes after insemination in the presence of 1 mM 3-aminotriazole (3AT; Sigma-Aldrich, St Louis, MO, USA). Excess sperm were separated from the fluid by centrifugation at 1000 g. The remaining supernatant was dialyzed against TE, then lyophilized and resuspended in distilled water; (5) soft fertilization envelopes (SFEs) were isolated from the washed zygotes used to collect zygotic seawater, as previously described (Wong and Wessel, 2004).
Polyclonal antibody analysis
One-dimensional polyacrylamide gel electrophoresis (PAGE) was accomplished in pre-cast 4-20% polyacrylamide Tris-Glycine gels (Nu-Sep Incorporated, Lawrenceville, GA, USA). Total protein was transferred onto high-capacity nitrocellulose (Pall Corporation, Pensacola, FL, USA), and then blocked using Blotto [3% non-fat dry milk (w/v), 170 mM NaCl, 50 mM Tris, 0.05% Tween20 (v/v) (pH 8)]. Primary antibodies were diluted in Blotto [eTG sera at 1:1000 dil or nTG sera at 1:000 dil (v/v)], followed by horseradish peroxidase-conjugated secondary antibodies diluted in Blotto [1:5000 dil (v/v); Jackson ImmunoResearch Laboratories Incorporated, West Grove, PA, USA]. Blots were washed extensively in TBS-Tween20 [170 mM NaCl, 50 mM Tris, 0.05% Tween20 (v/v) (pH 8)] before development. Specific proteins were detected by chemiluminescence using a luminol-hydrogen peroxide solution [1.25 mM luminol, 68 μM coumeric acid, 0.0093% hydrogen peroxide (v/v), 0.1 M Tris (pH 8)]. Following development, blots were each stripped with two washes in 200 mM glycine-HCl, 0.05% Tween20 (pH 2.5) at 80°C for 20 minutes, and then equilibrated to TBS-Tween20. Blots were then stained with 0.1% Amido Black dissolved in destain solution [10% acetic acid, 45% methanol (v/v)], followed by extensive destaining in the same solvent.
Immunofluorescence localization was performed on paraffin embedded tissue fixed with 4% paraformaldehyde. Antibody probing was performed essentially as described previously for double-labeling (Wessel and McClay, 1986). Anti-nTG sera [1:2000 dil (v/v)] was detected with anti-rabbit Cy5 Fab [1:25 dil (v/v); Jackson ImmunoResearch Laboratories Incorporated, West Grove, PA, USA], then remaining anti-nTG sites were blocked with unlabeled anti-rabbit Fabs [1:3 dil (v/v); Jackson ImmunoResearch Laboratories Incorporated, West Grove, PA, USA] (Negoescu et al., 1994). Hyalin staining was used as a counterstain for cortical granules, and was probed for at the same time as the unlabeled anti-rabbit Fab incubation. Anti-hyalin monoclonal antibody 2B7 [1:3 dil (v/v)] (Wessel et al., 1998) was detected with anti-mouse Alexa Fluor 564 (Invitrogen Corporation, Carlsbad, CA, USA). Finally, anti-eTG sera [1:100 dil (v/v)] was detected with anti-rabbit Alexa Fluor 488 IgGs (Invitrogen Corporation, Carlsbad, CA, USA). Samples were imaged using an LSM510 META laser confocal microscope (Carl Zeiss Corporation, Thornwood, NY, USA). Z-stacks were taken in 0.5 μm steps.
Electron microscopic analysis was performed on paraformaldehyde-fixed whole-mount eggs and zygotes (see above), immunoprobed with preimmune sera [1:200 dil (v/v)], anti-nTG sera [1:500 dil (v/v)], or anti-eTG sera [1:200 dil (v/v)] followed by anti-rabbit IgGs conjugated to 10 nm colloidal gold (Jackson ImmunoResearch Laboratories Incorporated, West Grove, PA, USA). These samples were prepared and imaged as previously described (Wong and Wessel, 2006b). Each colloidal gold particle was scored according to its location along the egg cortex or the fertilization envelope, and normalized over the length of each structure analyzed. Data are reported as the number of particles per millimeter, per egg or per embryo, averaged over three to six cells.
Inhibition studies: drug concentrations
All enzymatic inhibition assays were conducted on about 200 acid-dejellied eggs in a final volume of 200 μl of seawater. Unless otherwise noted, commercial inhibitors were used at a final concentration of: 10 mM cadaverine-HCl (cad; Sigma-Aldrich); 1× mini EDTA-free protease inhibitor (p.inh; Roche Diagnostics Corporation, Indianapolis, IN, USA); 10 μM diphenyleneiodonium (DPI; Sigma-Aldrich, St Louis, MO, USA); or 1 mM 3-aminotriazole (3AT; Sigma-Aldrich, St Louis, MO, USA).
In vivo cadaverine and tyramide labeling ± functional blocking analysis
Quantification of transglutaminase-dependent or peroxidase-mediated inter-protein crosslinking within the fertilization envelope was measured using the fluorophore conjugates cadaverine-Alexa Fluor 488 or tyramide-Alexa Fluor 594, respectively (Invitrogen Corporation, Carlsbad, CA, USA). About 200 acid-dejellied eggs were preincubated on ice for 30 minutes with various dilutions of purified IgGs (control used ASW only) or chemical inhibitors in 200 μl of seawater. An equal volume of 20 μM cadaverine-Alexa Fluor 488 and 1:4000 dilution of tyramide-Alexa Fluor 594 stock solution (see manufacturer handbook) were added with homospecific sperm (1:10,000 final concentration), diluting the IgGs (125, 250 or 500 pg affinity-purified IgG per egg) or chemical inhibitors to their final concentration (see the section `Inhibition studies'). Twenty minutes after insemination, zygotes were washed five times with ice-cold ASW, and then resuspended in a 1% paraformaldehyde solution in ASW. Incorporation of each competitor molecule was imaged on an LSM510 META laser confocal microscope (Carl Zeiss Incorporated, Thornwood, NY, USA) at the equatorial plane of the fertilization envelope. Fluorescence intensity in the fertilization envelope was quantified with Metamorph software (Molecular Devices, Sunnyvale, CA, USA). Mean values from 20 individual zygotes per treatment are reported.
Scanning electron microscopy
Morphological alterations of microvillar casts in the fertilization envelopes were observed on zygotes fertilized in the presence of respective inhibitors (250 pg/egg of each affinity purified anti-TG IgG was used), as above without Alexa Fluor conjugates. Zygotes were then fixed and post-processed as previously described (Battaglia and Shapiro, 1988). Samples were coated with gold-palladium (60:40) using an Emitech sputter coater (Emitech Limited, Kent, England) immediately before digital imaging on a Hitachi S-2700 scanning electron microscope (Hitachi High Technologies America, Pleasanton, CA, USA) using Quartz PCI software (Quartz Imaging Corporation, Vancouver, BC, Canada).
Chemical isolation of and analysis of labeled fertilization envelopes
Fertilization envelopes were chemically isolated from live zygotes inseminated in the presence or absence of 3-aminotriazole (Sigma-Aldrich, St Louis, MO, USA) as documented elsewhere (Santiago and Carroll, 1987; Veron et al., 1977). Briefly, 30 minutes after insemination in the presence of 100 μM cadaverine analogs [cadaverine HCl (cad-HCl) or cadaverine-Alexa Fluor 488 (cad-AF)], zygotes were washed three times with ASW and then twice with CFSW. Soft fertilization envelope were dissolved by incubating zygotes in 2 M urea, 100 mM DTT, 250 mM EDTA in CFSW for 15 minutes, and repeating once. Supernatants from successive incubations were pooled, and proteins were isolated using methanol chloroform precipitation (Wessel and Flugge, 1984) and resuspended in 2× sample buffer [5 mM Tris, 20% sucrose, 2% SDS (w/v) (pH 6.8)].
Polyacrylamide gel electrophoresis (PAGE) was accomplished in precast 4-20% polyacrylamide Tris-Glycine gels (Nu-Sep Incorporated, Lawrenceville, GA, USA) or using the ZOOM system (Invitrogen Corporation, Carlsbad, CA, USA). For the latter, fertilization envelope proteins were first focused on a linear ZOOM pH 3-10 isoelectric gradient and then subjected to separation on a 4-20% polyacrylamide Tris-Glycine gel (Invitrogen Corporation, Carlsbad, CA, USA). For both types of electrophoresis, gels were imaged for fluorescence using a Typhoon scanner (488 nm excitation, 526 band pass emission filter) driven by proprietary software (Amersham Biosciences, Piscataway, NJ, USA). Gels were then stained for total protein with Colloidal Coomassie (Nu-Sep Incorporated, Lawrenceville, GA, USA).
Mass spectrometry
In-gel samples were digested according to manufacturer instructions using a Pierce In-Gel Tryptic Digestion Kit (ThermoFisher Scientific Incorporated, Rockford, IL, USA), and then samples were cleaned using Pierce PepClean-18 columns (ThermoFisher Scientific Incorporated, Rockford, IL, USA). Resuspended peptides were processed on an LTQ Ion Trap mass spectrometer (ThermoFisher Scientific Incorporated, Rockford, IL, USA), and the spectra were analyzed with Bioworks and SEQUEST software programs (ThermoFisher Scientific Incorporated, Rockford, IL, USA).
Population-level measurement of hydrogen peroxide and ovoperoxidase activity
Hydrogen peroxide production or ovoperoxidase activity was measured for about 200 pretreated or control eggs using Amplex Red (Invitrogen Corporation, Carlsbad, CA, USA) on a microtiter plate system, as previously published (Wong et al., 2004). The output from each well was normalized to the kinetics of eggs or sperm alone, and is reported as percentage of control reactions (preimmune or ASW alone).
In vivo cadaverine incorporation on the egg surface by pH
pH dependence of transamidation on the egg surface was assessed using the crosslinking of cadaverine-Alexa Fluor 488 (Invitrogen Corporation, Carlsbad, CA, USA) to endogenous substrates in the vitelline layer or on the plasma membrane. About 100 freshly shed eggs were resuspended in seawater buffered with 10 mM citrate at various pH values (pH 5-8) to assess transglutaminase activation. To measure activity at a constant pH, the eggs were resuspended in 200 μl citrate-buffered ASW containing 10 μM cadaverine-Alexa Fluor 488. To measure activity in response to a rapid pH shift, the eggs were first incubated in the appropriate citrate-buffered ASW for 5 minutes, and then the fluid was exchanged for normal ASW (pH 8) containing 10 μM cadaverine-Alexa Fluor 488. Ten minutes after addition of cadaverine-Alexa Fluor 488 solutions, eggs were washed five times with ice-cold ASW, and then resuspended in a 1% paraformaldehyde solution in ASW. Conjugation of fluorophore to the egg surface was imaged on an LSM510 META laser confocal microscope (Carl Zeiss Incorporated, Thornwood, NY, USA), taking 20 μm z-stacks using 2-μm optical slices at 2-μm steps through the equatorial plane of the egg. Stacks were projected onto a single image for quantification using LSM510 software (Carl Zeiss Incorporated, Thornwood, NY, USA). Fluorescence intensity at the egg surface was quantified with Metamorph software (Molecular Devices, Sunnyvale, CA, USA). Mean values from five to eight individual eggs per treatment are reported.
Statistical analysis
Comparative analysis of cadaverine or tyramide conjugates within in tact fertilization envelopes was carried out using a two-tailed Student's t-test, comparing experimental sets against control only.
RESULTS
Two transglutaminases are differentially localized in the sea urchin oocyte and egg
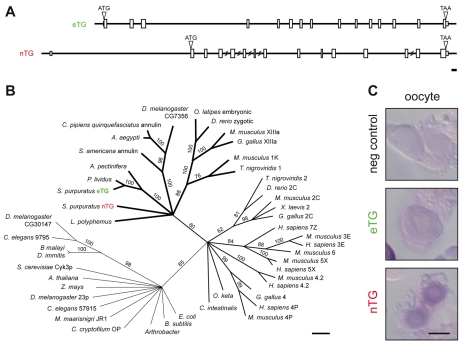
The genome of the sea urchin Strongylocentrotus purpuratus (Sea Urchin Genome Sequencing Consortium et al., 2006) contains two transglutaminases (TGs), named according to their subcellular location (as described below): an extracellular (eTG) and a nuclear (nTG) isoform. The two isozymes are derived from independent genes (Fig. 1A). Consistent with other family members (Lorand and Graham, 2003), neither S. purpuratus transglutaminase contains a signal peptide. Their open reading frames are 34% identical, with 26% and 29% identity between the N and C sequences outside of the core catalytic region, respectively (see Fig. S1 in the supplementary material). S. purpuratus eTG clusters with the two other echinoderm transglutaminases identified; both the eTG clade and S. purpuratus nTG are closely related to factor XIIIa and transglutaminase 1 of vertebrates (Fig. 1B).
Fig. 1.
Both Strongylocentrotus purpuratus transglutaminases are expressed in oocytes and eggs. (A) Scaled diagrams of each transglutaminase found in the S. purpuratus genome. Boxes indicate exons; ATG, predicted initiating methionine; TAA, putative translational stop codon. Scale bar represents 300 nucleotides, 100 amino acid residues. (B) Cladogram of the transglutaminase superfamily. Representative members include: [kingdom Monera] Arthrobacter species (ABK03932); Bacillus subtilis (CAB15105); Candidatus Korarchaeum cryptofilum OP (ACB07036); Escherichia coli (JC7310); Methanoculleus maarisnigri JR1 (ABN57069); [kingdom Fungi] Saccharomyces cerevisiae Cyk3p (Q07533); [kingdom Plantae] Arabidopsis thaliana (CAN87017); [kingdom Animalia] Aedes aegypti (EAT41371); Asterina pectinifera (BAB20439); Brugia malayi (AAQ23042); Caenorhabditis elegans 9795 (AAN09795), 57916 (CAB57916); Ciona intestinalis (CAA71263); Culex pipiens quinquefasciatus annulin (EDS39328); Danio rerio 2C (AAH78420), zgc (AAI24321); Dirofilaria immitis (AAC24752); Drosophila melanogaster CG30147-PA (AAF57449), CG7356-PA (AAF52590), RE12423p (AAN71312); Gallus gallus 2C (AAA49104), 4 (NP_001006368), XIIIa (CAC10657); Homo sapiens 3E (AAI09076), 4P (AAB95430), 4.2 (AAA74589), 5X (AAI22860), 7Z (NP_443187); Limulus polyphemus (2012342A); Mus musculus 1K (AAF35986), 2C (AAH16492), 3E (NP_033400), 4P (NP_808579), 4.2 (AAA62275), 5X (AAI16268), 6 (AAI12421), XIIIa (AAH40274); Oncorhynchus keta (BAA11633); Oryzias latipes embryonic (BAC84967); Paracentrotus lividus (CAD28789); Schistocerca americana annulin (P52183); Strongylocentrotus purpuratus eTG (FJ829470) and nTG (FJ829471); Tetraodon nigroviridis 1 (CAF97726), 2 (CAG03221); Xenopus laevis 2 (AAH56053). Numbers are bootstrap values from 1000 iterations representing heuristic searches performed using the default TBR branch-swapping algorithm of PAUP (Swofford, 2002). Scale equals 100 changes. (C) In situ RNA hybridization probing for each transglutaminase transcript in oocytes. Negative control probes for the exogenous transcript neomycin phosphotransferase II. Scale bar: 50 μm.
Each transglutaminase gene is expressed in the oocyte (Fig. 1C). Whereas eTG mRNA persists in eggs (1.13±0.06 units in eggs, relative to oocytes by quantitative PCR), nTG mRNA is largely degraded during meiotic maturation (0.31±0.03 units in eggs, relative to oocytes). Antibodies generated against each isozyme specifically identify the said proteins (see Fig. S1 in the supplementary material). Anti-nTG antiserum predominantly detects a protein of ∼110 kDa (Fig. 2A), near its predicted molecular weight (∼83.9 kDa). Anti-eTG serum detects a ∼210 kDa protein (Fig. 2A), a mass over twice the predicted size (∼82.9 kDa), suggesting eTG may exist as a homodimer that is stable under these analytical conditions. This association may occur during its secretion, as observed with other extracellular transglutaminases (Adany and Bardos, 2003; Griffin et al., 2002), because in vitro synthesis of eTG does not yield dimers (see Fig. S1C in the supplementary material).
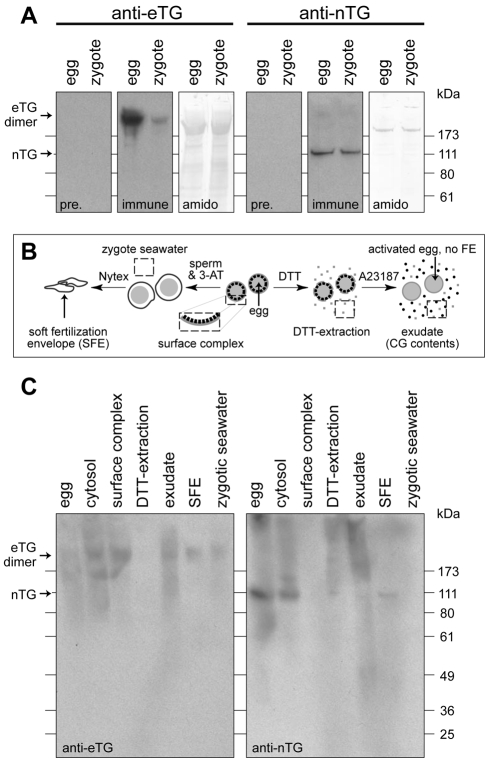
Fig. 2.
Transglutaminases are present inside and outside the cell. (A) Immunoblots for each transglutaminase in total egg or zygote extracts. Five hundred (∼20 μg total protein; anti-eTG) or 50 (∼2 μg total protein; anti-nTG) individual eggs or zygotes were probed per respective lane, and corresponding blots were stained for total protein using Amido Black. (B) Description of cell fractionation protocol. (C) Immunoblot for eTG and nTG in egg and zygote fractions, collected according to methods in B. Biochemical preparations tested are: egg (20 μg), cytosol (20 μg), surface complex (10 μg), exudate (10 μg), fertilization envelope (5 μg) and zygotic seawater (10 μg).
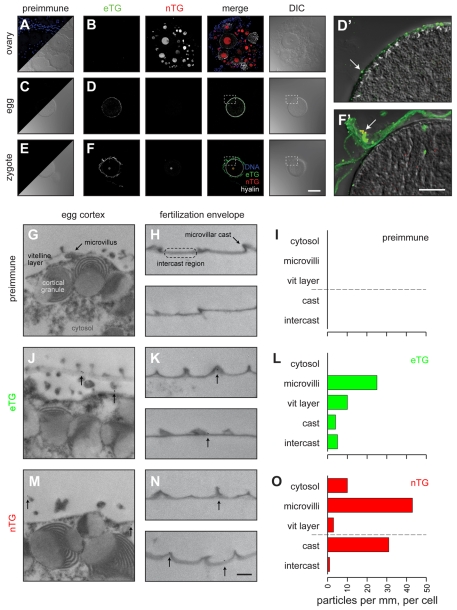
Biochemical fractionation and in situ localization also reveal differential distribution of the egg transglutaminases. nTG is highly enriched in the nucleus (Fig. 3B), although a fraction of the nTG population resides in the vitelline layer near the microvilli (Fig. 3L) and is retained in the fertilization envelope (Fig. 2C), primarily at the microvillar casts (Fig. 3F,L). eTG, conversely, is exclusively outside of the egg. It resides in the vitelline layer, enriched near microvilli (Fig, 3D,J,L), but is more evenly distributed throughout the mature fertilization envelope than the egg surface enrichment would predict (Fig. 3F,L). About 20% of the original egg population of eTG is retained with the zygote (Fig. 2A), while the remainder is probably released into the media (Fig. 2C).
Fig. 3.
Transglutaminases are enriched at egg microvilli and at microvillar casts in the fertilization envelope. (A-F) Immunofluorescence localization of each transglutaminase in sections of S. purpuratus ovary (A,B), eggs (C,D), zygotes (E,F) and fertilization envelopes (F′). Each image represents the projection of 10 μm optical stacks. Images from preimmune sera (A,C,E) are contrasted with antisera (B,D,F). Staining for each transglutaminase is shown in grayscale in their respective rows, and colored in the merged overlay. Sections were counterstained for the cortical granule/zygote extracellular matrix protein hyalin, as shown in white only in the merged images. Detailed magnification of egg (D') and zygote (F') are shown overlaid on the respective DIC images, with sources indicated by dashed boxes in parent image. Arrows indicate punctae that may represent microvilli (D') or microvillar casts (F'). Scale bars: 50 μm. (G-O) Immunogold labeling for each TG in egg cortex (G,J,M) and fertilization envelopes (H,K,N). Scale bar: 500 nm. (I,L,O) The number of particles found in specific subdomains (labeled in G,H) along the surface of the egg or the fertilization envelope. All data were transposed to a linear denominator (e.g. particles found within a 5 μm wide band of cytoplasm below the plasma membrane were `projected' onto a line equal to the length traced by the overlying membrane). Thus, the quantity of gold particles found within each subdomain is presented on a per millimeter, per cell basis in bar graphs for respective sera.
Both TGs participate in fertilization envelope transamidation
Native ε(γ-glutamyl)lysine crosslinking is difficult to detect because of the small mass change it adds to the participating proteins and the insolubility of the resultant aggregates (Nemes et al., 2005). To overcome these limitations, we used low concentrations of Alexa Fluor 488-conjugated cadaverine (cad-AF) as a reporter of in vivo transamidation (Fig. 4A). High concentrations of unlabeled cadaverine (cad-HCl) can inhibit protein crosslinking, just as with putrescine or glycine ethyl ester (Battaglia and Shapiro, 1988; Nemes et al., 2005). Thus, we used cadaverine analogs to overcome the two major hurdles to observing transglutaminase function in vivo.
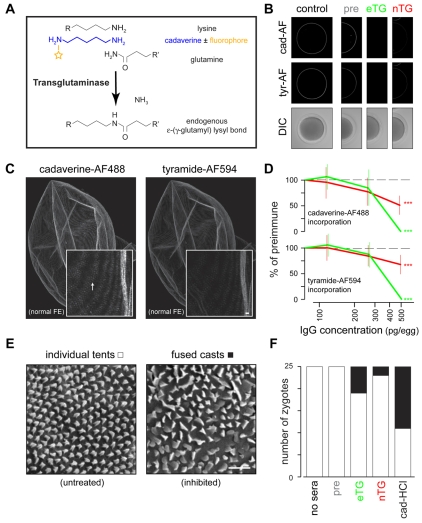
Fig. 4.
Transamidation is inhibited by functional blocking antibodies against transglutaminases. (A) Chemistry of endogenous transamidation, compared with chemistry with competitive substrate analogs of cadaverine. The fluorophore used in subsequent experiments is Alexa Fluor 488, e.g. cad-AF. (B) Representative images of cadaverine-Alexa Fluor 488 (cad-AF) and tyramide-Alexa Fluor 594 (tyr-AF) incorporation into the fertilization envelope. Zygotes shown were created in the presence of 0 (control) or 500 pg affinity-purified IgGs per egg; examples of the complete titration series can be found in Fig. S2 in the supplementary material. Respective fluorophore-reporters are shown compared with DIC images. Quantification of such labeling is shown in D. (C) Projected image from a normal fertilization envelope that incorporated cadaverine-Alexa Fluor 488 (left) and tyramide-Alexa Fluor 594 (right) reporters. This sample was mechanically separated from its respective zygote. Insets represent the same region of the fertilization envelope, in detail. Arrow indicates microvillar casts enriched with cadaverine-Alexa Fluor 488. Scale bar: 1 μm. (D) Quantification of equatorial cadaverine-Alexa Fluor 488 (top) or tyramide-Alexa Fluor 594 (bottom) incorporation into the fertilization envelope in the presence of specific affinity-purified anti-transglutaminase IgGs. All data are presented after normalization to respective preimmune IgG fluorescence. Standard deviations (vertical lines) from 20 zygotes per treatment are shown. ***P<0.0001. (E) Representative scanning electron micrographs of the surface of fertilization envelopes formed in the presence of preimmune IgGs (untreated) or anti-eTG IgGs (inhibited), showing zygotes possessing fertilization envelopes with individual or fused microvillar casts, respectively. Scale bar: 1 μm. (F) Frequency of zygotes exhibiting microvillar casts, as shown in E, per treatment. Fertilization envelopes were formed in the presence of 250 pg affinity-purified anti-transglutaminase IgGs per egg (pre, eTG, nTG) or 10 mM cadaverine-HCl, versus seawater controls.
Cadaverine analogs primarily affect the microvillar cast transition of the fertilization envelope in a dose-dependent fashion. When present during normal fertilization, cad-AF reporter accumulates at microvillar casts (Fig. 4C), consistent with the enriched transamidation in this subdomain of the fertilization envelope (Battaglia and Shapiro, 1988; Larabell and Chandler, 1991; Mozingo and Chandler, 1991). Inhibition of transamidation with excess cad-HCl disrupts the normal transmogrification of microvillar casts from an `igloo' to `tent' (I-to-T) shape (Battaglia and Shapiro, 1988; Larabell and Chandler, 1991; Mozingo and Chandler, 1991), leaving individual casts as `igloos' and/or promoting the fusion of neighboring casts (Fig. 4E). Inhibition of ovoperoxidase with 3-aminotriazole does not affect the I-to-T transition (Mozingo and Chandler, 1991), consistent with the relative exclusion of the dityrosine crosslinking reporter tyramide-Alexa Fluor 594 (Wong and Wessel, 2008) from the microvillar casts of normal fertilization envelopes (Fig. 4C).
Affinity-purified antibodies against both transglutaminases differentially inhibit extracellular transamidation and affect dityrosine crosslinking. Compared with preimmune IgG controls, functional blocking anti-TG IgGs block both cad-AF incorporation into the fertilization envelope (Fig. 4B,D) and the microvillar I-to-T transmogrification (Fig. 4E,F). Incorporation of tyramide-Alexa Fluor 594 was surprisingly sensitive to the loss of transamidation (Fig. 4B,D), consistent with previous observations that ovoperoxidase activity is affected by the TG inhibitor glycine ethyl ester (Foerder and Shapiro, 1977; Hall, 1978; Nemes et al., 2005; Turner et al., 1985).
Transamidation of specific substrates in the fertilization envelope
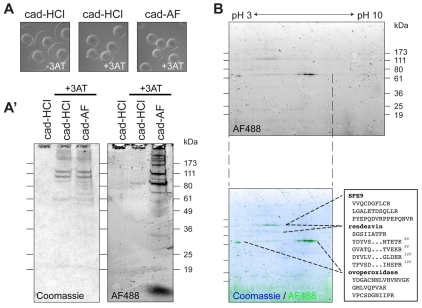
The covalent incorporation of the cad-AF reporter was used to identify transglutaminase substrates within the fertilization envelope. To enrich for labeled substrates, we dissolved fertilization envelopes with a solution of urea (Inoue and Hardy, 1971), dithiothreitol (Hall, 1978; Veron et al., 1977) and calcium chelators (Bryan, 1970; Carroll et al., 1986). Only softened fertilization envelopes formed in the presence of 3-aminotriazole (+3AT) were sensitive to these chemicals, as indicated by their solubility (Fig. 5A) and the migration of their constituents into a polyacrylamide gel (Fig. 5A′). Including low concentrations of cad-AF during fertilization envelope formation allowed us to identify specific proteins modified by transamidation (Fig. 5A′). Resolution of these tagged samples by two-dimensional electrophoresis, followed by mass spectrometry analysis of the resolved spots, identified SFE9, rendezvin and ovoperoxidase as TG substrates within the fertilization envelope (Fig. 5B).
Fig. 5.
Transamidation targets a subset of structural proteins and ovoperoxidase. (A) Zygotes following the chemical stripping procedure (see Materials and methods), showing that only fertilization envelopes formed in the presence of 3-aminotriazole (+3AT; no ovoperoxidase activity) are sensitive to chemical dissolution. The presence of cadaverine (cad-HCl) or cadaverine-Alexa Fluor 488 (cad-AF) does not alter this 3AT-dependent phenotype. (A′) One-dimensional SDS-PAGE of chemically dissolved fertilization envelopes collected from populations of zygotes in A. Equal volumes were loaded per lane, measuring ∼3.5 μg of total protein. Coomassie staining (left) and Alexa Fluor 488 fluorescence showing cad-AF incorporation (right) are shown. Autofluorescence at ∼80 kDa of the cad-HCl sample from +3AT samples is probably the heme group in ovoperoxidase (LaFleur et al., 1998). (B) Two-dimensional gel electrophoresis of ∼5 μg of chemically stripped fertilization envelopes formed in the presence of cad-AF. Isoelectric focusing (x-axis) was achieved on a linear pH 3-10 gradient. Fluorescence alone (top) and overlaid onto total protein (bottom; AF488 in green, Coomassie in blue) are shown. Inset is a sample of the peptide sequences identified from three major bands incorporating cadaverine Alexa Fluor 488. A complete list of the peptides can be found in Fig. S3 in the supplementary material.
Transglutaminases control dityrosine crosslinking through two paths
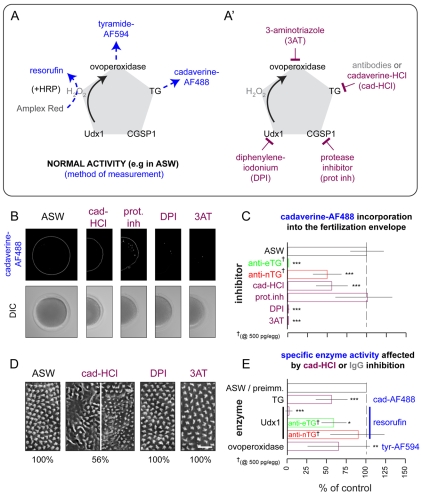
In addition to being transamidated (Fig. 5), ovoperoxidase is sensitive to transglutaminase (Fig. 4), CGSP1 (Haley and Wessel, 2004) and Udx1 (Wong et al., 2004) activity. Given this preliminary network of enzyme regulation, we investigated whether transamidation affects other enzymatic activities during fertilization envelope assembly. Reporters used to measure the major activities include (Fig. 6A): conversion of Amplex Red to resorufin by exogenous horseradish peroxidase to measure Udx1 production of hydrogen peroxide on a population level (Wong et al., 2004), or without horseradish peroxidase to measure ovoperoxidase activity (see Fig. S4 in the supplementary material); incorporation of tyramide-Alexa Fluor 594 to measure ovoperoxidase activity per zygote (Wong and Wessel, 2008); and incorporation of cad-AF to measure transamidation per zygote (Nemes et al., 2005).
Fig. 6.
Inter-relationship between transamidation and other fertilization envelope enzyme activities. (A) Schematic listing methods of measuring specific enzyme activities through fluorescent reporters. (A′) List of pharmacological inhibitors used to knock down specific enzymes used during fertilization envelope maturation. (B) Representative images of cadaverine-Alexa Fluor 488 incorporation into the fertilization envelope (top), after formation in the presence of the indicated chemical inhibitors, versus their respective DIC images (bottom). (C) Quantified results of cadaverine-Alexa Fluor 488 incorporation, when fertilization envelopes are formed in the presence of the indicated inhibitors (purple). All results are normalized to untreated controls (ASW). Anti-transglutaminase (green or red) results are from highest concentrations reported in Fig. 4. Standard deviations (lines) over at least four independent experiments representing 12 pools of 200 eggs (resorufin) or 20 individual fertilization envelopes are shown. 3AT and para-aminobenzoic acid produced similar results (data not shown). ***P<0.0001. (D) Representative scanning electron micrographs of fertilization envelope surfaces established in the presence of respective enzyme inhibitors. Frequency of the phenotype shown is indicated as a percentage of total zygotes scores (n=25); the remaining 44% of fertilization envelopes created in the presence of 10 mM cadaverine-HCl appear normal (e.g. ASW). (E) Quantification of enzyme activities obtained in the presence of 10 mM cadaverine-HCl (purple) or anti-TG IgGs (green or red) to block transamidation. Methods from A (blue) used to measure enzyme activity (black) are listed on the right. All results are normalized to controls (ASW or preimmune IgGs). Standard deviations (lines) for at least four independent experiments of representing 12 pools of 200 eggs (resorufin) or 20 individual fertilization envelopes are shown. *P=0.01; **P<0.001; ***P<0.0001. Complete datasets for C and E are presented in Fig. S4 in the supplementary material.
Inhibition of transamidation reduces both hydrogen peroxide production and ovoperoxidase activity. As expected, chemical or antibody inhibition of transamidation blocks cad-AF accumulation in the fertilization envelope (Fig. 6B,C). Excess cad-HCl and anti-eTG IgGs also suppress Udx1-dependent resorufin accumulation and ovoperoxidase activity (Fig. 6E; see Fig. S4 in the supplementary material). The specific knockdown of hydrogen peroxide production by anti-eTG might account for the extra potency of the anti-eTG versus anti-nTG on tyramide-Alexa Fluor 594 incorporation into the fertilization envelope (Fig. 4D; Fig. 6E), suggesting that both TGs control ovoperoxidase activity but eTG also regulates Udx1.
Environmental acidification activates extracellular transglutaminases
Analogous to its sibling factor XIII (Adany and Bardos, 2003; Esposito and Caputo, 2005), we predicted that proteolysis would activate the extracellular transglutaminases. Alternatively, oxidation by hydrogen peroxide could activate the catalytic cysteine residues in each TG (Griffin et al., 2002; Lorand and Graham, 2003). To distinguish these hypotheses, we assessed how the inhibition of each major enzyme activity affects cad-AF incorporation into the fertilization envelope and microvillar cast transmogrification (Fig. 6A′). We used protease inhibitors to block CGSP1 (Haley and Wessel, 1999), diphenyleneiodonium (DPI) to block Udx1 production of hydrogen peroxide (Wong et al., 2004) and 3-aminotriazole (3AT) to block ovoperoxidase (Showman and Foerder, 1979).
Surprisingly, Udx1 and ovoperoxidase, but not CGSP1, regulate fertilization envelope transamidation. Inhibition of either hydrogen peroxide production (DPI) or ovoperoxidase (3AT) blocks incorporation of cad-AF, whereas protease inhibitors have no direct effect (Fig. 6B,C). Protease inhibitors do, however, suppress hydrogen peroxide production by Udx1, and consequently deplete dityrosine crosslinking by ovoperoxidase (see Fig. S4 in the supplementary material). Consistent with previous observations (Carroll et al., 1986; Mozingo and Chandler, 1991), only direct inhibition of transamidation with excess cad-HCl affects the morphological transition of the microvillar casts (Fig. 6D). Thus, transamidation at the microvillar casts occurs before transglutaminase activity is modified by Udx1 or by ovoperoxidase (Fig. 6C).
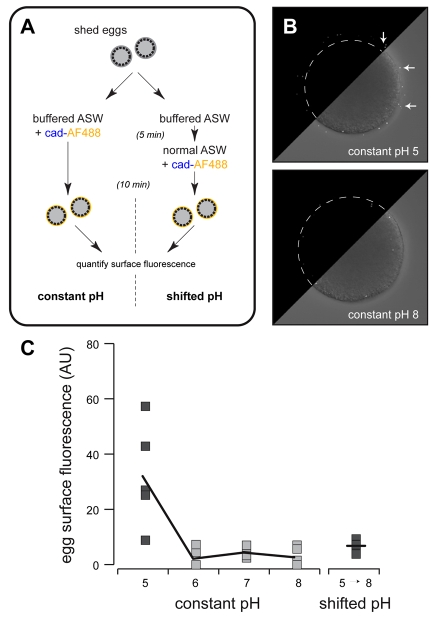
As CGSP1 does not affect transamidation, and transglutaminase activity occurs in advance of dityrosine crosslinking, we next sought non-enzymatic sources of transglutaminase activation. Many transglutaminases are activated by free Ca2+ (Lorand and Graham, 2003), but the 10 mM concentrations of Ca2+ in seawater are not sufficient to initiate transamidation. Instead, the activating factor is related to cortical changes during fertilization (Battaglia and Shapiro, 1988). The cell surface is transiently acidified between 30-120 seconds after fertilization (Paul et al., 1976; Smith et al., 2002), timing that is coordinate with exocytosis of cortical granule contents (Chandler and Heuser, 1979; Haley and Wessel, 2004; Kay and Shapiro, 1985; Matese et al., 1997) and with peak transamidation activity (Battaglia and Shapiro, 1988). Might acidic environments activate the transglutaminases, as observed for the teleost transglutaminase that crosslinks the chorion (Ha and Iuchi, 1998)? We tested this by determining whether cad-AF could be covalently linked to the egg surface by simply acidifying the seawater, mimicking the transient pH shift following fertilization (Fig. 7A). Indeed, lowering the seawater to pH to 5 increases transamidation (Fig. 7C), as reported by a punctate incorporation of cad-AF on the cell surface (Fig. 7B) that is consistent with the microvillar enrichment of these enzymes in eggs (Fig. 3). Surprisingly, a rapid shift from pH 5 to pH 8 by seawater exchange significantly abolished surface labeling (Fig. 7A,C). Thus, local acidification at the egg surface is sufficient to activate the transglutaminases, and the gradual return to alkaline pH 8 conditions 3-5 minutes after fertilization (Paul et al., 1976; Smith et al., 2002) probably inactivates them.
Fig. 7.
Acidic environments support transamidation. (A) Protocol used to test pH dependency of transamidation. Normal ASW is ∼pH 8. (B) Composite images represent individual eggs treated at pH 5 (top) and pH 8 (bottom). Upper left corner of each image shows fluorescence signal, whereas lower right corner is an overlay of fluorescence signal (white) over DIC image. Egg plasma membrane is outlined in fluorescence-only corners. Arrows indicate extracellular signal; most signal from pH 8 eggs is cytoplasmic. (C) Quantification of surface fluorescence in response to constant pH exposure or rapid alkalination from pH 5 to pH 8. Results from individual eggs are shown, with the averages per treatment connected by lines (n=5-8 eggs per pH, per condition). pH 5 results are significantly different than each other treatment (P≤0.01).
DISCUSSION
Identification of the transglutaminases (TGs) completes the molecular dissection of the sea urchin fertilization envelope, some 150 years since its first description (Derbès, 1847) and 100 years after its first major enzyme activity was measured (Warburg, 1908). Ironically, the last enzyme identified is the first one active during fertilization envelope assembly (Battaglia and Shapiro, 1988; Larabell and Chandler, 1991; Veron et al., 1977).
Both eTG and nTG reside on the egg surface until fertilization, when they are transiently activated and locally modify the fertilization envelope (Fig. 8). eTG is present as inactive homodimers, the most effective form for extracellular transglutaminases (Griffin et al., 2002) such as mammalian factor XIIIa (Adany and Bardos, 2003; Esposito and Caputo, 2005); nTG may be stored as a monomer or a heat-sensitive dimer. At fertilization, the acidic lumen of the cortical granule increases pH just prior to exocytosis (Morgan and Galione, 2007); these protons are pumped out of the cell, thereby temporarily acidifying the cell surface (Paul et al., 1976; Smith et al., 2002). This transient acidification reversibly isomerizes and activates the transglutaminases (Lorand and Graham, 2003); when the local environment returns to pH 8 after 3-5 minutes (Paul et al., 1976; Smith et al., 2002), transamidation is repressed (Battaglia and Shapiro, 1988). Transamidation could be further repressed by hydrogen peroxide-mediated oxidation of its catalytic cysteine (Lorand and Graham, 2003) and/or by excess dityrosine crosslinking (Foerder et al., 1978; Wong et al., 2004; Wong and Wessel, 2008) as both Udx1 and ovoperoxidase activities ramp up as transamidation peaks (Fig. 8).
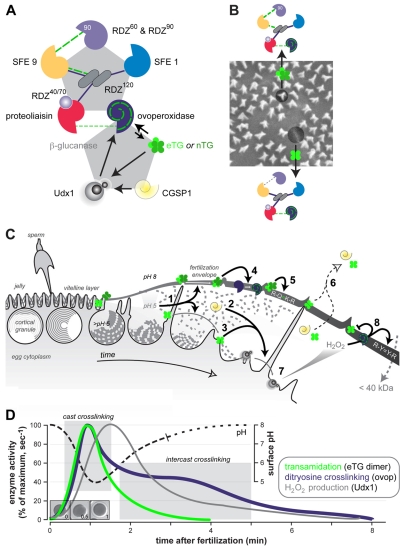
Fig. 8.
Schematic relationship between the proteome and enzome of the fertilization envelope. (A) The biochemical proteome, adapted, with permission, from Wong and Wessel (Wong and Wessel, 2006b), and its link to the enzyme regulatory network that is needed to assemble the fertilization envelope. No evidence implicates β-1,3-glucanase as a contributor to the formation or maturation of the fertilization envelope. Short broken lines indicate ionic protein binding; unbroken black lines indicate dityrosine crosslinking by ovoperoxidase; long broken green lines indicate transamidation by the transglutaminases; solid arrows indicate positive activation. (B) The binding proteome mapped onto the vitelline layer during fertilization envelope assembly. Protein symbols are as in A. (C) Chronology of enzyme activity during fertilization envelope assembly. Colorized enzymes are as in A. See text for details. Adapted, with permission, from Wong and Wessel (Wong and Wessel, 2008). (D) Time course of enzyme activities related to inter-protein crosslinking measured after fertilization occurs. Separate curves for transamidation (Battaglia and Shapiro, 1988), hydrogen peroxide production (Wong et al., 2004) and dityrosine crosslinking (Wong and Wessel, 2008) are taken or extrapolated from previous work, and are plotted against the surface pH changes measured previously (Smith et al., 2002). Images of eggs and zygotes are to temporal scale with the x-axis, showing that fertilization envelope assembly is completed 1 minute after fertilization. Shaded regions predict events that occur within the fertilization envelope at a given time.
We suggest the following network of extracellular modifying activities among the major enzymes contributing to fertilization envelope assembly (Fig. 8): coordinate with cortical granule exocytosis, local acidification at the egg surface (Paul et al., 1976; Smith et al., 2002) (1) activates both transglutaminases. Alkalinization of the cortical granule lumen (Morgan and Galione, 2007) primes the auto-activation of CGSP1 (Haley and Wessel, 1999) and ovoperoxidase (Deits and Shapiro, 1985) so they are active when secreted. CGSP1 activity (2) allows the egg vitelline layer to separate from the plasma membrane. Proteolysis also directly activates Udx1, perhaps releasing the N-terminal peroxidase domain from its NADPH reductase region (Wong et al., 2004) to lengthen the half-life of hydrogen peroxide. Within the first 2 minutes of fertilization (Larabell and Chandler, 1991), (3) a surface-associated population of eTG activates Udx1 while (4) the remaining transglutaminases are released with the vitelline layer. As cortical granule structural proteins are woven into the vitelline layer, active transglutaminases retained at the microvillar casts mediate the I-to-T transmogrification, crosslinking microvillar cast proteins such as SFE9 (Wessel, 1995) prior to any crosslinking of the intercast regions (Chandler and Heuser, 1980; Larabell and Chandler, 1991). Transglutaminases in the intercast region or in solution modify (or are modified by) active ovoperoxidase tethered within the fertilization envelope; the parallel profiles of transamidation and dityrosine crosslinking within the first 2 minutes of fertilization (Fig. 8D) support the coordinate regulation of these two enzymes. The initial phase of dityrosine crosslinking could, for example, correspond to transglutaminase-mediated regulation of ovoperoxidase at the microvillar casts (Mozingo et al., 1994): crosslinking could (5) lock ovoperoxidase in its active state following hysteresis (Deits et al., 1984; Deits and Shapiro, 1985), thereby reducing its sensitivity to peroxidase inhibitors and allowing it to complete crosslinking of the microvillar casts (Chandler and Heuser, 1980; Mozingo et al., 1994). Intercast ovoperoxidase could also be permanently tethered to proteoliaisin by transamidation, limiting its redistribution to other domains of the fertilization envelope (Mozingo et al., 1994; Weidman et al., 1985). (6) Soluble CGSP1 and active transglutaminases released from the zygote surface and/or not retained in the fertilization envelope both diffuse away from the zygote, while the permeability of the fertilization envelope remains unrestricted. (7) As production of hydrogen peroxide from Udx1 increases, (8) ovoperoxidase-dependent crosslinking increases among the structural proteins. This burst of dityrosine crosslinking may also covalently modify the transglutaminases retained in the fertilization envelope, permanently shutting them off. This leaves only proteoliaisin-tethered ovoperoxidase to complete inter-protein crosslinking (Chandler and Heuser, 1980; Larabell and Chandler, 1991; Mozingo et al., 1994), thereby establishing the permeability barrier associated with the mature fertilization envelope (Kay and Shapiro, 1985; Veron et al., 1977; Wong and Wessel, 2008).
This regulatory enzome of the sea urchin fertilization envelope is parsimonious with decades of observations. First, blocking transamidation impairs microvillar cast transformation (Battaglia and Shapiro, 1988; Chandler and Kazilek, 1986; Mozingo and Chandler, 1991), but also reduces total protein content (Kay and Shapiro, 1985; Mozingo and Chandler, 1991) and overall thickness (Cariello et al., 1994), refractivity (Cariello et al., 1994) and permeability (Kay and Shapiro, 1985; Veron et al., 1977) of the fertilization envelope - characteristics that have been attributed primarily to dityrosine crosslinking. Second, the transglutaminase inhibitor glycine ethyl ester (Nemes et al., 2005) blocks ovoperoxidase activity at high concentrations (Foerder and Shapiro, 1977; Hall, 1978; Turner et al., 1985). Third, despite the activity of transglutaminase-dependent crosslinking, a single peroxidase inhibitor [e.g. diphenyleneiodonium (Wong et al., 2004), 3-aminotriazole (Showman and Foerder, 1979) or para-aminobenzoic acid (Hall, 1978)] is sufficient to abolish crosslinking of the intercast regions of the fertilization envelope (Mozingo et al., 1994), making it sensitive to mechanical shearing, chemical dissolution and gel electrophoresis (Showman and Foerder, 1979; Wessel, 1995; Wong et al., 2004). Finally, inhibition of CGSP1 blocks inter-protein crosslinking (Carroll et al., 1986) associated with thickening of the microvillar casts and intercast regions (Mozingo and Chandler, 1991). The same inhibitor also blocks hydrogen peroxide production (Coburn et al., 1981) and, logically, establishment of the fertilization envelope permeability barrier (Epel et al., 1970; Wong and Wessel, 2008).
Based on the knowledge that cadaverine analogs are conjugated to endogenous glutamine residues (Nemes et al., 2005), we predict the following transamidation pairings within the fertilization envelope: (1) SFE9 and the vitelline layer rendezvin120 may stabilize the microvillar casts, consistent with the enrichment of SFE9 within this subdomain (Wessel, 1995) and the ability to incorporate cadaverine-Alexa Fluor 488 to the vitelline layer at the egg surface (Fig. 7). (2) Transamidation may tether specific proteins to the fertilization envelope. Ovoperoxidase glutamines, for example, could be covalently bound to proteoliaisin lysines, thereby establishing more permanent association between these two proteins (Somers et al., 1989; Weidman et al., 1985); it is still possible to resolve the two proteins after pharmacological inhibition of ovoperoxidase (Deits et al., 1984; Showman and Foerder, 1979) because transamidation is repressed in the intercast region, where the majority of these proteins reside in the expanded fertilization envelope (Mozingo et al., 1994). Alternatively, (3) crosslinking of intra-enzyme glutamine and lysine residues may positively regulate ovoperoxidase, perhaps locking it in an active state following hysteretic modification (Deits and Shapiro, 1985; Deits and Shapiro, 1986). We hypothesize that a glutamine-conjugated analog would produce results consistent with these substrate pairings.
Thus, the two major crosslinking activities required for sea urchin fertilization envelope assembly are co-regulated. The major structural proteins of the S. purpuratus fertilization envelope proteome (Wong and Wessel, 2006b) are crosslinked by these enzymes at either microvillar casts or intercast domains (Wong and Wessel, 2008), establishing a mechanically `hardened' shell for early embryogenesis. The relatively uniform contribution of these two enzyme activities is conserved among sea urchins (Cariello et al., 1994; Veron et al., 1977), even though other animals do not require two enzymes to achieve an analogously `hardened' matrix: Transamidation is sufficient to harden the matrices of mosquito (Iijima et al., 2005) and most teleost eggs (Chang et al., 2002; Ha and Iuchi, 1998; Oppen-Berntsen et al., 1990; Yamagami et al., 1992); peroxidase is required for the teleost Tribulon (Kudo, 1988); and a zinc-dependent protease accomplishes the same task for amphibians (Lindsay and Hedrick, 2004). What, then, do these complementary interactions imply about the assembly of the sea urchin fertilization envelope? Might transamidation activity be carried over from its role during extracellular matrix modifications in the oocyte?
Post-translational modification is essential for the regulation of protein activity and is capable of changing individual molecules and/or cellular morphology. When these modifications are absent or go awry, however, the associated changes can gravely affect tissue integrity such that overall health is at risk. The absence of transamidation negatively impacts the survival of some animal zygotes (Battaglia and Shapiro, 1988; Chandler and Kazilek, 1986; Chang et al., 2002; Ha and Iuchi, 1998; Larabell and Chandler, 1991; Mozingo and Chandler, 1991; Oppen-Berntsen et al., 1990; Veron et al., 1977; Yamagami et al., 1992). Similarly, inhibition of an extracellular transglutaminase blocks normal peroxidase-dependent isodityrosine crosslinking of Chlamydomonas reinhardtii cell walls, which is lethal to the single cell alga (Waffenschmidt et al., 1999; Waffenschmidt et al., 1993). Aberrant transglutaminase activity is also associated with human pathologies, including skin defects (Candi et al., 2005; Lorand and Graham, 2003), blood clotting disorders (Adany and Bardos, 2003; Lorand and Graham, 2003) and neurodegenerative diseases (Aeschlimann and Thomazy, 2000; Cooper et al., 2002). Similarly, excess NADPH oxidase or peroxidase activity, each of which generates reactive oxygen radicals, can accelerate oxidative damage of proteins generally (Davies et al., 1987a; Davies et al., 1987b; Fridovich, 1998), expand the severity of tissue damage at sites of bacterial infection (Heinecke, 1999; Winterbourn et al., 2000) or at athleroschlerotic plaques (Jacob et al., 1996; Lambeth, 2007), advance the aging process through disruption of tissue integrity (Levine and Stadtman, 2001), and/or facilitate the proliferation of cancer (Lambeth, 2007). Reduction of peroxidase activity, via loss of peroxidase or its source of hydrogen peroxide, is also associated with vasodilation (Wang et al., 1993), congenital hypothyroidism (Bakker et al., 2000; Moreno et al., 2002) and increased susceptibility to infection (Lambeth, 2007; Winterbourn et al., 2000). The intersection between transamidation, dityrosine crosslinking and disease is clearest in the immune system, where misregulation of the orchestrated enzyme network could domino into chronic inflammation owing to improper clotting (Lorand and Graham, 2003) or excessive oxidative damage (Heinecke, 1999; Lambeth, 2007; Winterbourn et al., 2000). Although directed gene expression is a crucial source of regulation on a cellular level, additional levels of control are essential once these enzymes are released into the extracellular matrix. Our current knowledge that sea urchin transglutaminases and ovoperoxidase regulate each other during fertilization envelope assembly may reveal how other extracellular matrix modifiers are managed throughout development and homeostasis, allowing us to differentiate normal from pathological activities.
Supplementary material
Supplementary material available online at http://dev.biologists.org/cgi/content/full/136/11/1835/DC1
Supplementary Material
We thank the Providence Institute of Molecular Oogenesis, including Lianne Davis and Anthony Morgan, and Alison De Long for her constructive criticism. This work was funded by the NSF and NIH. Deposited in PMC for release after 12 months.
References
- Adany, R. and Bardos, H. (2003). Factor XIII subunit A as an intracellular transglutaminase. Cell Mol. Life Sci. 60, 1049-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschlimann, D. and Thomazy, V. (2000). Protein crosslinking in assembly and remodelling of extracellular matrices: the role of transglutaminases. Connect. Tissue Res. 41, 1-27. [DOI] [PubMed] [Google Scholar]
- Antonyak, M. A., Jansen, J. M., Miller, A. M., Ly, T. K., Endo, M. and Cerione, R. A. (2006). Two isoforms of tissue transglutaminase mediate opposing cellular fates. Proc. Natl. Acad. Sci. USA 103, 18609-18614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Mena, C., Cameron, A. R. and Davidson, E. H. (2000). Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development 127, 4631-4643. [DOI] [PubMed] [Google Scholar]
- Bakker, B., Bikker, H., Vulsma, T., de Randamie, J. S., Wiedijk, B. M. and De Vijlder, J. J. (2000). Two decades of screening for congenital hypothyroidism in The Netherlands: TPO gene mutations in total iodide organification defects (an update). J. Clin. Endocrinol. Metab. 85, 3708-3712. [DOI] [PubMed] [Google Scholar]
- Battaglia, D. E. and Shapiro, B. M. (1988). Hierarchies of protein cross-linking in the extracellular matrix: involvement of an egg surface transglutaminase in early stages of fertilization envelope assembly. J. Cell Biol. 107, 2447-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, J. (1970). On the reconstitution of the crystalline components of the sea urchin fertilization membrane. J. Cell Biol. 45, 606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi, E., Schmidt, R. and Melino, G. (2005). The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328-340. [DOI] [PubMed] [Google Scholar]
- Cariello, L., Zanetti, L. and Lorand, L. (1994). Effects of inhibiting transglutaminase during egg fertilization and development. Biochem. Biophys. Res. Commun. 205, 565-569. [DOI] [PubMed] [Google Scholar]
- Carroll, D. J., Acevedo-Duncan, M., Justice, R. W. and Santiago, L. (1986). Structure, assembly and function of the surface envelope (fertilization envelope) from eggs of the sea urchins, Strongylocentrotus purpuratus. Adv. Exp. Med. Biol. 207, 261-291. [DOI] [PubMed] [Google Scholar]
- Carroll, E. J., Jr and Epel, D. (1975). Isolation and biological activity of the proteases released by sea urchin eggs following fertilization. Dev. Biol. 44, 22-32. [DOI] [PubMed] [Google Scholar]
- Carroll, E. J., Jr, Byrd, E. W. and Epel, D. (1977). A novel procedure for obtaining denuded sea urchin eggs and observations on the role of the vitelline layer in sperm reception and egg activation. Exp. Cell Res. 108, 365-374. [DOI] [PubMed] [Google Scholar]
- Chandler, D. E. and Heuser, J. (1979). Membrane fusion during secretion: cortical granule exocytosis in sea urchin eggs as studied by quick-freezing and freeze-fracture. J. Cell Biol. 83, 91-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, D. E. and Heuser, J. (1980). The vitelline layer of the sea urchin egg and its modification during fertilization: a freeze-fracture study using quick-freezing and deep-etching. J. Cell Biol. 84, 618-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, D. E. and Kazilek, C. J. (1986). Extracellular coats on the surface of Strongylocentrotus purpuratus eggs: stereo electron microscopy of quick-frozen and deep-etched specimens. Cell Tissue Res. 246, 153-161. [DOI] [PubMed] [Google Scholar]
- Chang, Y. S., Wang, Y. W. and Huang, F. L. (2002). Cross-linking of ZP2 and ZP3 by transglutaminase is required for the formation of the outer layer of fertilization envelope of carp egg. Mol. Reprod. Dev. 63, 237-244. [DOI] [PubMed] [Google Scholar]
- Chen, M. Y., Hu, K. Y., Huang, C. C. and Song, Y. L. (2005). More than one type of transglutaminase in invertebrates? A second type of transglutaminase is involved in shrimp coagulation. Dev. Comp. Immunol. 29, 1003-1016. [DOI] [PubMed] [Google Scholar]
- Coburn, M., Schuel, H. and Troll, W. (1981). A hydrogen peroxide block to polyspermy in the sea urchin Arbacia punctulata. Dev. Biol. 84, 235-238. [DOI] [PubMed] [Google Scholar]
- Cooper, A. J., Jeitner, T. M., Gentile, V. and Blass, J. P. (2002). Cross linking of polyglutamine domains catalyzed by tissue transglutaminase is greatly favored with pathological-length repeats: does transglutaminase activity play a role in (CAG)(n)/Q(n)-expansion diseases? Neurochem. Int. 40, 53-67. [DOI] [PubMed] [Google Scholar]
- Davies, K. J., Delsignore, M. E. and Lin, S. W. (1987a). Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J. Biol. Chem. 262, 9902-9907. [PubMed] [Google Scholar]
- Davies, K. J., Lin, S. W. and Pacifici, R. E. (1987b). Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J. Biol. Chem. 262, 9914-9920. [PubMed] [Google Scholar]
- Deits, T. and Shapiro, B. M. (1985). pH-induced hysteretic transitions of ovoperoxidase. J. Biol. Chem. 260, 7882-7888. [PubMed] [Google Scholar]
- Deits, T. L. and Shapiro, B. M. (1986). Conformational control of ovoperoxidase catalysis in the sea urchin fertilization membrane. J. Biol. Chem. 261, 12159-12165. [PubMed] [Google Scholar]
- Deits, T., Farrance, M., Kay, E. S., Medill, L., Turner, E. E., Weidman, P. J. and Shapiro, B. M. (1984). Purification and properties of ovoperoxidase, the enzyme responsible for hardening the fertilization membrane of the sea urchin egg. J. Biol. Chem. 259, 13525-13533. [PubMed] [Google Scholar]
- Derbès, A. (1847). Observations sur le mecanisme et les phenomenes qui accompagnent la formation de l'embryon chezl'oursin comestible. Ann. Sci. Nat. Zool. 8, 80-98. [Google Scholar]
- Detering, N. K., Decker, G. L., Schmell, E. D. and Lennarz, W. J. (1977). Isolation and characterization of plasma membrane-associated cortical granules from sea urchin eggs. J. Cell Biol. 75, 899-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel, D., Weaver, A. M. and Mazia, D. (1970). Methods for removal of the vitelline membrane of sea urchin eggs. I. Use of dithiothreitol (Cleland Reagent). Exp. Cell Res. 61, 64-68. [DOI] [PubMed] [Google Scholar]
- Esposito, C. and Caputo, I. (2005). Mammalian transglutaminases: identification of substrates as a key to physiological function and physiopathological relevance. FEBS J. 272, 615-631. [DOI] [PubMed] [Google Scholar]
- Foerder, C. A. and Shapiro, B. M. (1977). Release of ovoperoxidase from sea urchin eggs hardens the fertilization membrane with tyrosine crosslinks. Proc. Natl. Acad. Sci. USA 74, 4214-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerder, C. A., Klebanoff, S. J. and Shapiro, B. M. (1978). Hydrogen peroxide production, chemiluminescence, and the respiratory burst of fertilization: interrelated events in early sea urchin development. Proc. Natl. Acad. Sci. USA 75, 3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich, I. (1998). Oxygen toxicity: a radical explanation. J. Exp. Biol. 201, 1203-1209. [DOI] [PubMed] [Google Scholar]
- Griffin, M., Casadio, R. and Bergamini, C. M. (2002). Transglutaminases: nature's biological glues. Biochem. J. 368, 377-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C. R. and Iuchi, I. (1998). Enzyme responsible for egg envelope (chorion) hardening in fish: purification and partial characterization of two transglutaminases associated with their substrate, unfertilized egg chorion, of the rainbow trout, Oncorhynchus mykiss. J. Biochem. (Tokyo) 124, 917-926. [DOI] [PubMed] [Google Scholar]
- Haley, S. A. and Wessel, G. M. (1999). The cortical granule serine protease CGSP1 of the sea urchin, Strongylocentrotus purpuratus, is autocatalytic and contains a low-density lipoprotein receptor-like domain. Dev. Biol. 211, 1-10. [DOI] [PubMed] [Google Scholar]
- Haley, S. A. and Wessel, G. M. (2004). Regulated proteolysis by cortical granule serine protease 1 at fertilization. Mol. Biol. Cell 15, 2084-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, H. G. (1978). Hardening of the sea urchin fertilization envelope by peroxidase-catalyzed phenolic coupling of tyrosines. Cell 15, 343-355. [DOI] [PubMed] [Google Scholar]
- Heinecke, J. W. (1999). Mass spectrometric quantification of amino acid oxidation products in proteins: insights into pathways that promote LDL oxidation in the human artery wall. FASEB J. 13, 1113-1120. [DOI] [PubMed] [Google Scholar]
- Iijima, M., Hashimoto, T., Matsuda, Y., Nagai, T., Yamano, Y., Ichi, T., Osaki, T. and Kawabata, S. (2005). Comprehensive sequence analysis of horseshoe crab cuticular proteins and their involvement in transglutaminase-dependent cross-linking. FEBS J. 272, 4774-4786. [DOI] [PubMed] [Google Scholar]
- Inoue, S. and Hardy, J. P. (1971). Fine structure of the fertilization membranes of sea urchin embryos. Exp. Cell Res. 68, 259-272. [DOI] [PubMed] [Google Scholar]
- Jacob, J. S., Cistola, D. P., Hsu, F. F., Muzaffar, S., Mueller, D. M., Hazen, S. L. and Heinecke, J. W. (1996). Human phagocytes employ the myeloperoxidase-hydrogen peroxide system to synthesize dityrosine, trityrosine, pulcherosine, and isodityrosine by a tyrosyl radical-dependent pathway. J. Biol. Chem. 271, 19950-19956. [DOI] [PubMed] [Google Scholar]
- Karlsson, C., Korayem, A. M., Scherfer, C., Loseva, O., Dushay, M. S. and Theopold, U. (2004). Proteomic analysis of the Drosophila larval hemolymph clot. J. Biol. Chem. 279, 52033-52041. [DOI] [PubMed] [Google Scholar]
- Kay, E. S. and Shapiro, B. M. (1985). The formation of the fertilization membrane of the sea urchin egg. In Biology of Fertilization, vol. 3 (ed. C. B. Metz and A. Monroy), pp. 45-80. Orlando, FL: Acadmic Press. [Google Scholar]
- Kay, E. S. and Shapiro, B. M. (1987). Ovoperoxidase assembly into the sea urchin fertilization envelope and dityrosine crosslinking. Dev. Biol. 121, 325-334. [DOI] [PubMed] [Google Scholar]
- Kinsey, W. H. (1986). Purification and properties of the egg plasma membrane. In Methods in Cell Biology, vol. 27 (ed. T. E. Schroeder), pp. 139-152. Orlando, FL: Academic Press. [DOI] [PubMed] [Google Scholar]
- Kudo, S. (1988). Chorionic peroxidase activity in the eggs of the fish Tribolodon hakonesis. J. Exp. Zool. 245, 63-70. [Google Scholar]
- LaFleur, G. J., Jr, Horiuchi, Y. and Wessel, G. M. (1998). Sea urchin ovoperoxidase: oocyte-specific member of a heme-dependent peroxidase superfamily that functions in the block to polyspermy. Mech. Dev. 70, 77-89. [DOI] [PubMed] [Google Scholar]
- Lambeth, J. D. (2007). Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radic. Biol. Med. 43, 332-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larabell, C. and Chandler, D. E. (1991). Fertilization-induced changes in the vitelline envelope of echinoderm and amphibian eggs: self-assembly of an extracellular matrix. J. Electron Microsc. Tech. 17, 294-318. [DOI] [PubMed] [Google Scholar]
- Levine, R. L. and Stadtman, E. R. (2001). Oxidative modification of proteins during aging. Exp. Gerontol. 36, 1495-1502. [DOI] [PubMed] [Google Scholar]
- Lindsay, L. L. and Hedrick, J. L. (2004). Proteolysis of Xenopus laevis egg envelope ZPA triggers envelope hardening. Biochem. Biophys. Res. Commun. 324, 648-654. [DOI] [PubMed] [Google Scholar]
- Lorand, L. and Graham, R. M. (2003). Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140-156. [DOI] [PubMed] [Google Scholar]
- Matese, J. C., Black, S. and McClay, D. R. (1997). Regulated exocytosis and sequential construction of the extracellular matrix surrounding the sea urchin zygote. Dev. Biol. 186, 16-26. [DOI] [PubMed] [Google Scholar]
- Mhaouty-Kodja, S. (2004). Ghalpha/tissue transglutaminase 2, an emerging G protein in signal transduction. Biol. Cell 96, 363-367. [DOI] [PubMed] [Google Scholar]
- Mian, S., el Alaoui, S., Lawry, J., Gentile, V., Davies, P. J. and Griffin, M. (1995). The importance of the GTP-binding protein tissue transglutaminase in the regulation of cell cycle progression. FEBS Lett. 370, 27-31. [DOI] [PubMed] [Google Scholar]
- Moreno, J. C., Bikker, H., Kempers, M. J., van Trotsenburg, A. S., Baas, F., de Vijlder, J. J., Vulsma, T. and Ris-Stalpers, C. (2002). Inactivating mutations in the gene for thyroid oxidase 2 (THOX2) and congenital hypothyroidism. N. Engl. J. Med. 347, 95-102. [DOI] [PubMed] [Google Scholar]
- Morgan, A. J. and Galione, A. (2007). Fertilization and nicotinic acid adenine dinucleotide phosphate induce pH changes in acidic Ca(2+) stores in sea urchin eggs. J. Biol. Chem. 282, 37730-37737. [DOI] [PubMed] [Google Scholar]
- Mozingo, N. M. and Chandler, D. E. (1991). Evidence for the existence of two assembly domains within the sea urchin fertilization envelope. Dev. Biol. 146, 148-157. [DOI] [PubMed] [Google Scholar]
- Mozingo, N. M., Somers, C. E. and Chandler, D. E. (1994). Ultrastructure of the proteoliaisin-ovoperoxidase complex and its spatial organization within the Strongylocentrotus purpuratus fertilization envelope. J. Cell Sci. 107, 2769-2777. [DOI] [PubMed] [Google Scholar]
- Nagahara, H., Vocero-Akbani, A. M., Snyder, E. L., Ho, A., Latham, D. G., Lissy, N. A., Becker-Hapak, M., Ezhevsky, S. A. and Dowdy, S. F. (1998). Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat. Med. 4, 1449-1452. [DOI] [PubMed] [Google Scholar]
- Negoescu, A., Labat-Moleur, F., Lorimier, P., Lamarcq, L., Guillermet, C., Chambaz, E. and Brambilla, E. (1994). F(ab) secondary antibodies: a general method for double immunolabeling with primary antisera from the same species: efficiency control by chemiluminescence. J. Histochem. Cytochem. 42, 433-437. [DOI] [PubMed] [Google Scholar]
- Nemes, Z., Petrovski, G., Csosz, E. and Fesus, L. (2005). Structure-function relationships of transglutaminases-a contemporary view. Prog. Exp. Tumor Res. 38, 19-36. [DOI] [PubMed] [Google Scholar]
- Oppen-Berntsen, D. O., Helvik, J. V. and Walther, B. T. (1990). The major structural proteins of cod (Gadus morhua) eggshells and protein crosslinking during teleost egg hardening. Dev. Biol. 137, 258-265. [DOI] [PubMed] [Google Scholar]
- Paul, M., Johnson, J. D. and Epel, D. (1976). Fertilization acid of sea urchin eggs is not a consequence of cortical granule exocytosis. J. Exp. Zool. 197, 127-133. [DOI] [PubMed] [Google Scholar]
- Piacentini, M., Rodolfo, C., Farrace, M. G. and Autuori, F. (2000). `Tissue' transglutaminase in animal development. Int. J. Dev. Biol. 44, 655-662. [PubMed] [Google Scholar]
- Santiago, L. and Carroll, E. J., Jr (1987). Intermolecular cross-linking of vitelline envelope polypeptides predominates in the hardened sea urchin fertilization envelope. Gamete Res. 17, 63-75. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium, Sodergren, E., Weinstock, G. M., Davidson, E. H., Cameron, R. A., Gibbs, R. A., Angerer, R. C., Angerer, L. M., Arnone, M. I., Burgess, D. R. et al. (2006). The genome of the sea urchin strongylocentrotus purpuratus. Science 314, 941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showman, R. M. and Foerder, C. A. (1979). Removal of the fertilization membrane of sea urchin embryos employing aminotriazole. Exp. Cell Res. 120, 253-255. [DOI] [PubMed] [Google Scholar]
- Smith, R. M., Baibakov, B., Lambert, N. A. and Vogel, S. S. (2002). Low pH inhibits compensatory endocytosis at a step between depolarization and calcium influx. Traffic 3, 397-406. [DOI] [PubMed] [Google Scholar]
- Somers, C. E., Battaglia, D. E. and Shapiro, B. M. (1989). Localization and developmental fate of ovoperoxidase and proteoliaisin, two proteins involved in fertilization envelope assembly. Dev. Biol. 131, 226-235. [DOI] [PubMed] [Google Scholar]
- Song, J. L., Wong, J. L. and Wessel, G. M. (2006). Oogenesis: single cell development and differentiation. Dev. Biol. 300, 385-405. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods). Sunderland, MA: Sinauer Associates.
- Turner, E., Somers, C. E. and Shapiro, B. M. (1985). The relationship between a novel NAD(P)H oxidase activity of ovoperoxidase and the CN- -resistant respiratory burst that follows fertilization of sea urchin eggs. J. Biol. Chem. 260, 13163-13171. [PubMed] [Google Scholar]
- Verderio, E. A., Johnson, T. and Griffin, M. (2004). Tissue transglutaminase in normal and abnormal wound healing: review article. Amino Acids 26, 387-404. [DOI] [PubMed] [Google Scholar]
- Veron, M., Foerder, C., Eddy, E. M. and Shapiro. (1977). Sequential biochemical and morphological events during assembly of the fertilization membrane of the sea urchin. Cell 10, 321-328. [DOI] [PubMed] [Google Scholar]
- Waffenschmidt, S., Woessner, J. P., Beer, K. and Goodenough, U. W. (1993). Isodityrosine cross-linking mediates insolubilization of cell walls in Chlamydomonas. Plant Cell 5, 809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt, S., Kusch, T. and Woessner, J. P. (1999). A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol. 121, 1003-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. X., Poon, C. I., Poon, K. S. and Pang, C. C. (1993). Inhibitory actions of diphenyleneiodonium on endothelium-dependent vasodilatations in vitro and in vivo. Br. J. Pharmacol. 110, 1232-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg, O. (1908). Beobachtungen uber die Oxydationsprozesse im Seeigelei. Z. Physiol. Chem. 57, 1-16. [Google Scholar]
- Weidman, P. J., Kay, E. S. and Shapiro, B. M. (1985). Assembly of the sea urchin fertilization membrane: isolation of proteoliaisin, a calcium-dependent ovoperoxidase binding protein. J. Cell Biol. 100, 938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, D. and Flugge, U. I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141-143. [DOI] [PubMed] [Google Scholar]
- Wessel, G. M. (1995). A protein of the sea urchin cortical granules is targeted to the fertilization envelope and contains an LDL-receptor-like motif. Dev. Biol. 167, 388-397. [DOI] [PubMed] [Google Scholar]
- Wessel, G. M. and McClay, D. R. (1986). Two embryonic, tissue-specific molecules identified by a double-label immunofluorescence technique for monoclonal antibodies. J. Histochem. Cytochem. 34, 703-706. [DOI] [PubMed] [Google Scholar]
- Wessel, G. M., Berg, L., Adelson, D. L., Cannon, G. and McClay, D. R. (1998). A molecular analysis of hyalin-a substrate for cell adhesion in the hyaline layer of the sea urchin embryo. Dev. Biol. 193, 115-126. [DOI] [PubMed] [Google Scholar]
- Winterbourn, C. C., Vissers, M. C. and Kettle, A. J. (2000). Myeloperoxidase. Curr. Opin. Hematol. 7, 53-58. [DOI] [PubMed] [Google Scholar]
- Wong, J. L. and Wessel, G. M. (2004). Major components of a sea urchin block to polyspermy are structurally and functionally conserved. Evol. Dev. 6, 134-153. [DOI] [PubMed] [Google Scholar]
- Wong, J. L. and Wessel, G. M. (2006a). Defending the zygote: search for the ancestral animal block to polyspermy. Curr. Top. Dev. Biol. 72, 1-151. [DOI] [PubMed] [Google Scholar]
- Wong, J. L. and Wessel, G. M. (2006b). Rendezvin: an essential gene encoding independent, differentially-secreted egg proteins that organize the fertilization envelope proteome following self-association. Mol. Biol. Cell 17, 5241-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J. L. and Wessel, G. M. (2008). Free-radical crosslinking of specific proteins alters the function of the egg extracellular matrix at fertilization. Development 135, 431-440. [DOI] [PubMed] [Google Scholar]
- Wong, J. L., Créton, R. and Wessel, G. M. (2004). The oxidative burst at fertilization is dependent upon activation of the dual oxidase Udx1. Dev. Cell 7, 801-814. [DOI] [PubMed] [Google Scholar]
- Wong, J. L., Koppel, D. E., Cowan, A. E. and Wessel, G. M. (2007). Membrane hemifusion is a stable intermediate of exocytosis. Dev. Cell 12, 653-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami, K., Hamazaki, T. S., Yasumasu, S., Masuda, K. and Iuchi, I. (1992). Molecular and cellular basis of formation, hardening, and breakdown of the egg envelope in fish. Int. Rev. Cytol. 136, 51-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.