Fig. 1.
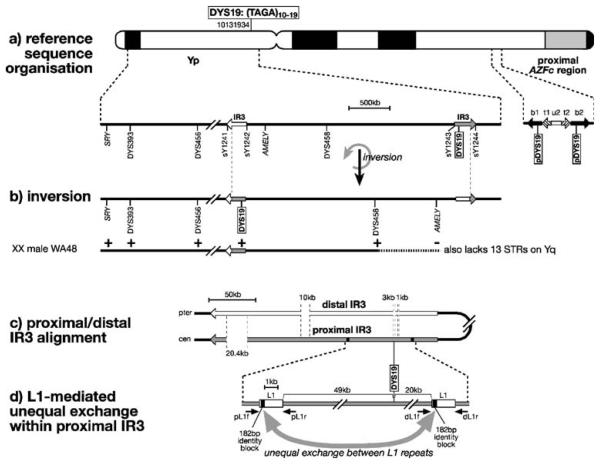
Complex genomic environment of DYS19 and putative mechanisms for inversions, deletions and duplications. a Reference sequence organisation around DYS19, showing position of the Y-STR on an idiogram of the Y chromosome (with genome position of start of the marker given according to build 36.1 of the reference assembly), and below, a schematic view of the region around DYS19 and its paralogues (pDYS19) on Yq. Arrows indicate inverted repeats (IR3 elements) or large repeat units in the region proximal to AZFc [42]. STSs and other markers used in mapping are shown below. b Structure of Yp following IR3-mediated inversion, with breakpoints indicated by dotted lines and mapping of presence (+) or absence (−) of markers in XX male WA48. The horizontal dotted line indicates the region of uncertainty of the breakpoint, resulting from the wide marker spacing. c Alignment of the IR3 inverted repeats; in the reference sequence organisation, DYS19 lies within the proximal IR3, corresponding to a 3-kb gap in the distal IR3. d Putative mechanism of DYS19 deletion or duplication mediated by unequal exchange (curved grey arrow) between flanking direct partial L1 repeats within the proximal IR3. Small arrows indicate PCR primers used to seek junction PCR products in deletion and duplication chromosomes