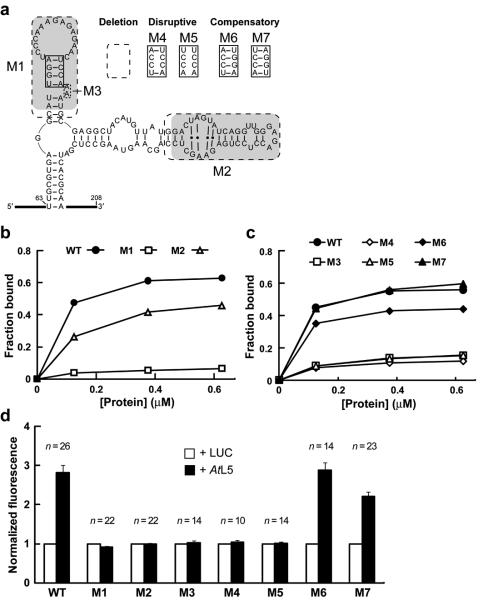
Figure 5.
The P2 stem of P5SM RNA is critical for binding to GST-AtL5 in vitro and disruption of AtL5 binding abolishes regulation of splicing in vivo. (a) Sequences of wild type (WT), deletion mutants, and P2 stem mutants of P5SM RNA used to assess domain contributions to protein binding. Shaded regions have sequence and structural similarity to corresponding shaded regions in 5S rRNA (Fig. 1d). Dashed boxes indicate nucleotides removed in deletion constructs (M1 through M3). The solid boxed region in the P2 stem was subjected to the disruptive (M4, M5) and compensatory (M6, M7) mutations shown. (b, c) Representative graphs of in vitro binding analyses for WT and mutant P5SM RNAs with different concentrations (0-0.625 μM) of GST-AtL5 protein. In c, the same symbol designates a disrupted mutant (open symbols) and its corresponding compensation mutant (filled symbols). The data lines for M3 and M5 are overlapping. (d) In vivo expression analysis for Pre-EGFP reporter constructs with co-expression of AtL5 in N. benthamiana. Pre-EGFP variants incorporate mutations M1 through M7 in the P5SM element. Data for the Pre-EGFP WT construct from Fig. 3c is shown for reference. For each construct, the EGFP fluorescence measured with expression of luciferase (LUC) was set to a value of 1 to enable comparison of relative change in response to AtL5 overexpression. This normalization procedure does not allow comparison of relative expression levels between the P5SM constructs, which instead were measured as shown in Supplementary Fig 9. Numbers of independent leaf samples (n) measured are shown. Error bars represent SEM.