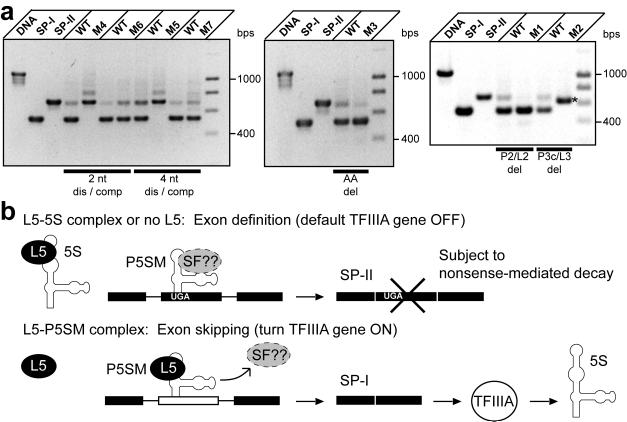
Figure 6.
The constitutive splicing patterns of deregulated mutants reveal that P5SM is involved in both exon definition and skipping, leading to a proposed model for regulation of TFIIIA pre-mRNA splicing. (a) RT-PCR detection of splice products arising from splicing of WT versus mutant Pre-EGFP reporter constructs. Each set consists of WT and one mutant reporter construct transformed on half of the same leaf to ensure near identical conditions for comparison. Effects from varying endogenous L5 levels were minimized by analyzing splice ratios upon constitutive AtL5 expression. Reporter fluorescence for these constructs measured under the same conditions for many individual leaves are shown in Supplementary Fig. 9. Total RNA was isolated from the same tissue amount for each leaf half, and roughly 1 μg was used in the RT reactions. Bars define disruptive (dis) and compensatory (comp) mutant pairs or deletion (del) mutants. Also shown are PCR products corresponding to unspliced precursor, SP-IE, and SP-IIE derived from DNA templates. The major splice product for the M2 mutant (labeled with an asterisk) is an SP-II type product which is shorter than the normal SP-II because of deleted sequence in the construct. DNA markers (M) from 400 to 1000 are in 200 bps increments. (b) The P5SM-L5 complex activates exon skipping, leading to the splice product (SP-I) that encodes full-length TFIIIA, whereas the default splice product (SP-II) is a nonsense-mediated decay substrate. Evidence suggests that P5SM is important in both exon recognition and skipping events. In the proposed mechanism, L5 displaces a putative exon-defining splice factor (SF) from P5SM. Thus, the discovery and analysis of this structured RNA element has elucidated a role for ribosomal protein L5 in the regulation of 5S rRNA synthesis in plants.