Abstract
In order to describe a cell at molecular level, a notion of a “gene” is neither necessary nor helpful. It is sufficient to consider the molecules (i.e., chromosomes, transcripts, proteins) and their interactions to describe cellular processes. The downside of the resulting high resolution is that it becomes very tedious to address features on the organismal and phenotypic levels with a language based on molecular terms. Looking for the missing link between biological disciplines dealing with different levels of biological organization, we suggest to return to the original intent behind the term “gene”. To this end, we propose to investigate whether a useful notion of “gene” can be constructed based on an underlying notion of function, and whether this can serve as the necessary link and embed the various distinct gene concepts of biological (sub)disciplines in a coherent theoretical framework. In reply to the Genon Theory recently put forward by Klaus Scherrer and Jürgen Jost in this journal, we shall discuss a general approach to assess a gene definition that should then be tested for its expressiveness and potential cross-disciplinary relevance.
Introduction
In a recent issue of this journal, Klaus Scherrer and Jürgen Jost (Scherrer and Jost 2007b) introduced an essentially computational account of gene expression, which introduces a formal separation of the “gene” from the program that is required to orchestrate its expression.
The Genon theory presents a fresh and stimulating contribution to a discussion of the “gene concept” that has re-emerged in recent years in response to evidence of greater genomic complexity than previous concepts of the gene are able to accommodate. It has become increasingly obvious that the classical molecular concept of a gene as a contiguous stretch of DNA encoding a functional product is inconsistent with the complexity and diversity of genomic organization (The ENCODE Project Consortium 2007; Maeda et al. 2006; Carninci 2006; Willingham and Gingeras 2006). Many of the proposals from the “high-throughput community” lean towards a purely structural point of view, focusing on genes as structural units, often explicitly related to proteins as the link to a functional interpretation (Snyder and Gerstein 2003; Gerstein et al. 2007). Dissenting opinions, on the other hand, question the usefulness of “genes” in genomic context (Gerstein et al. 2007).
The Genon theory attempts to reconcile these views by advocating a functional, rather than structural, definition of the gene. While this is a welcome departure from the overly simplistic view of “genes as protein-coding DNA”, it remains oriented toward the simple representation of the “gene” as a contiguous stretch of code. It deliberately excludes the complex collection of regulatory signals from the notion of the “gene” and instead interprets them as a program of gene expression, the “genon”. It is grounded in a number of fundamental assumptions, some implicit and some explicit. Our discussion will start with these assumptions, which in several case are not satisfying. Instead of presenting a particular fixed definition of what a gene “is”, we will explore here how a functional gene definition can be constructed depending on how the concept of “function” is formalized.
Gene expression as computation
The dichotomy of gene (data) and genon (program) is a fundamental assumption regarding the nature of biological information processing that is logically suspicious. In Computer Science, many of the familiar programming languages, including C, BASIC, or FORTRAN, make a clear syntactic distinction between data and program; functional programming languages such as LISP and Haskell, on the other hand, have no means at all for making this distinction. Since heritable biological information necessarily must encode both data and program, it is by no means clear that biological information processing is more like FORTRAN than LISP.
As an alternative to the separation into genes and genons, a separation into genetic material (data) and the machinery (program) that orchestrates its expression could be introduced. The latter respects an important intuitive property of data, namely the simple transfer and substitution of (parts of) the data. Similar to the platform-independence of data—in contrast to often platform-dependent programs—nucleic acids can be interpreted in a wide range of contexts. Biotechnology, and cloning techniques in general (Sambrook and Russel 2001), take advantage of this property whenever a piece of genetic material is cloned into a vector and transferred to a different organism. There a different machinery evaluates the same sequence information and generates a product that is similar enough to the original context to be of practical use.
Notwithstanding the appealing intuition behind this distinction, RNA components of the machinery inherited by an RNA molecule (as in the case of RNA viruses) pose a problem to this separation, because the same molecule would be both data and program at the same time. Therefore, it remains to be shown that an unambiguous partitioning of the molecular components into data and program is possible and that it results in a reasonable representation of biological reality.
A central idea of Genon Theory is that one can speak of a program that governs the expression of a gene. This program is described as the union of the cis-genon, which is encoded by the same molecule(s) that carry the information of the gene, and the trans-genon. The latter is viewed as the collection of all “trans-acting” factors that influence gene expression. The implicit assumption here is that the expression of the gene of interest does not change its environment in an appreciable manner, e.g., by using up some of the trans-factors or by feeding back on the expression of these factors. Only in this limiting case does it make sense to view the environment as a static part of the expression program, i.e., to associate the trans-genon with the gene of interest, instead of interpreting the environment, including the relevant trans-acting factors, as the result of other programs that concurrently express their genes. This static view of a set of “trans-acting” factors also fails to account for the fact that the expression of these factors is a dynamic process and will typically not be in sync with the processing steps of the gene of interest. We argue that specifying the collection of trans-acting factors is insufficient to determine the “external” part of the program of gene expression because the temporal order in which they are produced and interact is crucial.
Scherrer and Jost pre-suppose several properties of the process of gene expression. It is assumed to be deterministic (at least under given environmental conditions), Markovian (in the sense that each processing step only requires the result of the previous step as input), and to proceed in a linear sequence of a few well-separated steps. Each of these assumptions is an idealization. The last two properties together are necessary to justify the “Cascade of Regulation” and to make the notions of pre-genon, proto-genon, etc. well-defined. As the authors themselves note in (Scherrer and Jost 2007a), this assumption is often violated. Recent evidence for a strong coupling for transcription, splicing, and export in higher eukaryotes (Listerman et al. 2006; Swinburne et al. 2006; Maciag et al. 2006), and the concurrency of transcription and translation in bacterial cells (Gowrishankar and Harinarayanan 2004; El- Sharoud and Graumann 2007) implies that some of the processing stages may never exist as discrete molecules. This blurs the boundaries between the individual steps.
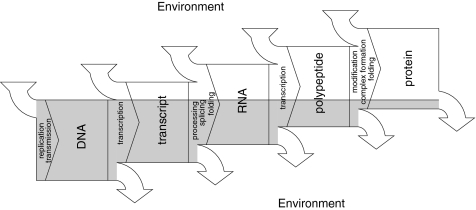
The separation of processing steps is, however, required to strictly distinguish cis- and trans-parts of the genon. Whenever a processing step results in joining two fragments (e.g., in trans-splicing), the element in trans becomes a cis-element after completing the step. The Markov property is also violated by splicing and some export mechanisms that specifically attach proteins that remain bound to the RNA during the next maturation step(s). Again it becomes impossible to strictly discriminate between cis- and trans-action. Exon-junction complexes and export co-factors such as the RNA binding protein HuR are of course not encoded in the final mRNA, but regulation of the mRNA depends on their presence and location in the pre-mRNA. This “annotation” is not seen in the final mRNA molecule, but is determined by the molecule’s particular processing history (Fig. 1).
Fig. 1.
Cascade of regulation: At each step, information content is not only reduced but might also increase due to integration of information provided by the surrounding. Global environmental factors (e.g., gravity, latitude, temperature, tide etc.) as well as local environmental factors including localization, timing and interaction of products provide information to all steps of the cascade and establish a network of communication. The influence of certain factors can be expected to show great variation among organisms. Localization is suggested to play an important role for many steps. The more environmental factors can be taken for granted, the less information needs to be encoded and transmitted from step to step
The Genon theory describes gene expression as a simple sequential program, thereby ignoring the network structure of gene regulation. In our view; however, the network architecture is the very essence of biological regulation. Within a framework that interprets gene expression as a computational process, we suggest reformulation of the trans-genon as communication with other gene expression processes. This leads in a rather natural way to a picture of gene expression as a distributed computing system (Attiya and Welsh 2004). To this end, we must give up the idea that there is a single, independent program governing the expression of each individual gene (one mRNA/gene–one genon hypothesis). Instead, we need to model a collection of computational processes—one for each sequence of consecutive processing steps—that communicate via their trans-actions. Formal models of this type have recently been introduced in systems biology (Danos and Laneve 2004; Danos et al. 2007; Kuttler and Niehren 2006) using π-calculus and related formalisms.
Genes sensu Jost and Scherrer
The Genon Theory emphasizes a functional point of view and attempts to define the gene as a “basis of a unit function”.1 It deliberately “give[s] up the correspondence of the gene as functional unit and as a DNA locus”. While there are rules to map genes back to the genome, these rules are not considered a defining property of the gene. Heritability, on the other hand, is. Jost and Scherrer, though, seem to view heritability as irrelevant, arguing that modern molecular biology is essentially about function.
We strongly disagree with this view. The concept of the “Gene” is common ground to most disciplines of biology and historically has been instrumental in the synthesis of subdisciplines, e.g., evolution and development. We therefore argue that a meaningful notion of “Gene” cannot be constructed with only a particular sub-discipline in mind. Heritability is a crucial property since it is the purpose of genomes to transmit the encoded instructions for generating functional units, instead of transmitting the functional units themselves. Even within the scope of modern molecular biology, the concept of heritable genes is indispensable: we need to be able to speak of homology—most commonly defined as descent from a common ancestor—among genes. Common ancestry of functional units is the main justification for translational approaches that attempt to utilize information obtained for model organisms such as mouse or fruitfly to understand similar biological processes in humans. Furthermore, it appears that genes are necessary to understand the selection part of the evolutionary process: In order to describe what selection does on a molecular level, only nucleotide sequences are required; to conceptualize the why, however, a functionally defined gene is at least very useful.
Scherrer and Jost proceed to equate function with “functional products” derived from the genetic encoding: “A cellular function can be represented by a polypeptide or an RNA”, “Genetic function is carried out by proteins composed of folded polypeptides”. Despite a section on RNA genes, the text leaves no doubt that protein-coding genes are considered the paradigm of genetic information processing; indeed, the Genon Theory fails to provide concepts to incorporate non-protein-coding “genes” in general. A more implicit assumption of the Genon Theory is the idea that protein coding mRNAs are the most interesting and most important type of products that are produced from DNA. In light of the results of the ENCODE and FANTOM projects we reject this “proteinocentric” point of view. Protein-coding sequence covers less than 2% of the genome, while approximately 10% is under stabilizing selection. This is at least indicative of some biological function. As almost all of this sequence is transcribed we have to assume that much of it exerts its function as some processing product of the primary transcript, which is often not associated with any protein (Pheasant and Mattick 2007). From this point of view, nothing about the mature mRNA stage is so special as to warrant the definition of this stage, along with the regulation of translation, as the focal point of biological information processing.
From these assumptions, Scherrer and Jost deduce that there is a single stage in the life of a transcript that lends itself to a natural definition of the gene, namely the last processing product before translation: “[The gene] finally emerges as an uninterrupted nucleic acid sequence at mRNA level, just prior to translation, in faithful correspondence with the amino acid sequence to be produced as a polypeptide”. The gene concept thus coincides with the well-established notion of “Open Reading Frame”. Consequently, there are many more (protein-coding) genes than protein coding loci (the authors estimate 500,000 vs. 25,000), since any two mRNAs giving rise to distinct polypeptides (e.g., via alternative splicing) are counted as distinct genes. On the other hand, the expression of the same function (i.e., the same functional molecule) at different times or in different cells counts as a single gene.
It is overly restrictive, however, to identify cellular functions with directly encoded gene products. Several classes of important molecules, all of which are “functional” (at least to most researchers), including steroid hormones, co-enzymes, pigments, polysaccharides, etc., are not directly encoded, but are quite indirectly the consequence of genetic encoding. Conversely, the polypeptide that is obtained directly by decoding the mRNA is in many cases not functional at all. It may need the assistance of chaperons to fold into its active tertiary structure, it may need to be modified, e.g., by glycosylation or other chemical modification, or it may be cleaved or fused with other (possibly modified) peptide chains. More importantly, there are crucial regulatory functions in which a process, e.g., the act of transcription to modify the chromatin state (Shearwin et al. 2005; Mazo et al. 2007), or the act of initial translation to remove the exon–junction complexes (Isken and Maquat 2007), is crucial, while the associated products created by these processes (a primary transcript and a polypeptide, respectively) are completely irrelevant for all we know.
On the other hand, function need not be associated with the generation of a product at all, as is the case with cis-acting regulatory elements. A classical example is the lac operator lacO (Jacob and Monod 1961). Besides cis dominance, this sequence shows properties similar to a regulatory gene and can be mapped to a DNA locus by means of physical mapping just like a gene. The Genon Theory thus uses a notion of “genetic” function that appears to be inconsistent with the experimental evidence.
Structural gene definitions
Less than 15 years ago, the influential textbook Genes V (Lewin 1994) defined: “Gene (cistron) is the segment of DNA involved in producing a polypeptide chain; it includes regions preceding and following the coding region (leader and trailer) as well as intervening sequences (introns) between individual coding segments (exons).” Older definitions explicitly included promoters as part of the gene. Once it had been realized; however, that the regulatory sequence associated with gene expression can be widely dispersed, many authors opted for viewing the “gene” as essentially synonymous to “protein-coding transcript” (Snyder and Gerstein 2003).
With the availability of large amounts of “omics” data, many authors have advocated various versions of structural definitions of the gene that amount to collections of transcripts, see, e.g., (Snyder and Gerstein 2003; Gerstein et al. 2007). The same approach is taken by current genome databases: within the 2 framework, a gene is defined as a set of (primary) transcripts. It seems that the gene definition of Scherrer and Jost was also influenced by this trend: even though introduced as a functional notion, a series of simplifying assumptions reduce it to another easily identifiable genomic structure: the Open Reading Frame.
A purely structural definition of a gene in terms of a genomic “source”, however, does not seem useful to us. Without any reference to function, there is no way of singling out a particular product of the regulatory cascade in general or a specific processing stage of a transcript in particular. As the end-product of every transcript is eventually a small degradation fragment, and presumably a single nucleotide, this approach does not lead to a meaningful definition. Alternatively, one might view every processing stage as a different transcript and consequently as a different gene. This would just rename “transcript” to “gene” and the set of all genes would become equivalent to the transcriptome. Another approach is to define a gene as a collection of overlapping transcripts. At least in eukaryotes, this leads to fairly large regions equivalent to genomic/transcriptional domains or, in the worst case, the whole genome, another trivial solution. Between these two extremes, Gerstein et al. (2007) consider genes as sets of overlapping transcripts that share open reading frames. As we have argued above, singling out particular processing stages or products is problematic since such a definition can be applied only to a (possibly small) subset of entities.
Genes derived from heritable functional units
We agree with Scherrer and Jost that a meaningful definition of gene has to be based on a notion of function because a purely structural gene definition is altogether dispensable as we have seen above. In this section, we will briefly outline a research agenda that may eventually lead to a useful function-based gene concept—or to the realization that such an endeavor cannot succeed.
First, we reject the idea of a one-to-one correspondence of function and “gene-product”, which seems much more a vestige of the history of the gene concept than a property of a biological system. The appeal of the equivalence of function and product is that it makes function “measurable” by virtue of detecting the product. We have argued above, however, that the existence of a product does not imply that it has any function at all, and conversely, the same product may have multiple and mechanistically diverse biochemical functions, depending on its context.
Hence, we expand the notion of function and postulate that function must be measurable directly by some experimental setup in finite time, and that one must be able to do this in such a way that functional equivalence can be determined. What constitutes a function, and whether two functions are distinguishable from each other, therefore depends on an experimental (or computational) procedure, which we will for short call a “measurement” in the following. Different procedures may represent “biological importance” more or less well. Time-honored procedures such as the classical complementation test of molecular genetics or the observation of the developmental effects of gene knock-outs are procedures that have proven useful. The approach of the Genon Theory, namely to determine whether a stretch of DNA is eventually translated into a polypeptide is yet another possible way to measure. We view computational approaches as yet another procedure to assess information about function. Of course, as with any “functional test”, all these procedures come with inherent limitations and the possibility of false positive and negative results. Such results may eventually lead to erroneous conclusions about particular “genes”. This is, however, also true for seemingly straightforward procedures such as the assignment of ORFs (Brent 2005), and does not affect the conceptual framework.
Entire cells, organs, and organisms certainly convey function. Thus we would not want to be forced to call everything that has a measurable function a “gene”. Just as Scherrer & Jost do, we consider a gene a unit of function. The nature of units, modules and their mutual relationships is a field of lively debate in theoretical biology, see, e.g., (Kvasnicka and Pospıchal 2002; Tanaka et al. 2006; Schlosser 2002; Wagner et al. 2007), which we will not enter here. Instead, we use the term “unit” in a broad sense: a unit should show stronger cohesion to itself than to other components, thereby ensuring its integrity in isolation. Consequently, a unit of function should execute its function in isolation, 3 thereby representing a “building block” or “basis element” of the space of functions. 4 Novel functions may emerge from collections of functional sub-units. Within a given experimental protocol we may be able to distinguish the function of higher level units from those of their components, thus functional units can be nested within each other. Intuitively, we would like to correlate the gene with the elementary functional unit, i.e., a unit that cannot be understood as a collection of functional units together with the emergent function(s) arising from their combination. Whereas single molecules and/or molecular complexes and their interactions play the central role in molecular biology, researchers in other biological disciplines might be more interested in higher order functional units. Such a coarse-grained level of functionality could be represented by chemical reactions, interaction networks, or phenotypic traits rather than products as functional units. We suggest that each of these is a valid starting point for a gene definition.
In contrast to the Genon Theory, we postulate that genes are heritable and therefore need to be part of the inherited material. In 1952, Hershey and Chase found that the “instructions” for functional units are made of genetic material, nucleic acid in general, DNA if present. However, exceptions to this rule are well known, e.g., epigenes, protein-based inheritance (i.e., centriols and prions) and RNA-based inheritance (Lolle et al. 2005) do instruct heritable functional units. Heritability is determined by the process of inheritance, a sequence of reproduction and segregation. We may or may not want to restrict the concept of genes to entities that are inherited in a particular way, namely by means of the genetic material that comprises the genome.
A formal mathematical investigation of this schema should eventually be able to relate elementary functional units to their source in the inherited material. If a function-based gene concept is feasible at all, such a mapping is the indispensable pre-requisite for genes to become a useful notion for molecular biology. We suspect that such a mapping is not necessarily possible for all underlying definitions of “function”, “unit” and/or their combinations. It is even conceivable that such a mapping can never be constructed, in which case we will have to abandon the notion of “functional genes”. Even if we can construct the map, there is no guarantee that the genomic source 5 corresponding to a particular definition of functional unit will show properties that we would expect or desire from a gene. In particular, the genomic representation of our functionally defined genes may well be frustratingly complex and disparate from the physical entities that we deal with in the various flavors of “omics”.
In line with our arguments above we suggest that an appropriate definition of a functional unit should not make explicit reference to a particular class of molecules. While determining the chemical composition is within the scope of acceptable experimental protocols, a consequence of this type of protocol is the disparate classification of molecules with similar or identical functions, e.g., a protein enzyme versus a ribozyme that catalyzes the same chemical reaction. It is at least conceivable that the chemical implementation of a catalyst or regulator is irrelevant for a cell. Consequently, functional units may just as well be of DNA nature. Operators and other cis-regulatory elements behave much like regulatory genes when assayed with many procedures typically used in genetics. In such a context, we may well be obliged to treat them as functional units and consequently as genes. On the other hand, Developmentally Regulated DNA Rearrangements (DRDR) are not uncommon as mechanisms of expression regulation throughout eukaryotes (Zufall et al. 2005). Ciliate genome processing (which interestingly is regulated by small RNAs (Garnier et al. 2004)), chromatin diminution (i.e., the selective elimination of portions of chromosomes), the vertebrate immune system, and the amplification of rDNA genes are the most prominent examples. DRDR is also involved in mating type switching in yeast and prokaryotic differentiation, see, e.g., (Carrasco et al. 1995). Hence processes operating on the genomic material have to be included in the processing program.
The boundaries of our genes as Heritable Elementary Functional Units are eventually determined by the underlying notion of function. Depending on this choice, genes may or may not contain the information necessary to orchestrate the production of the corresponding functional units from the heritable material.
Concluding remarks
In our discussion, we started from assumptions similar to but less restrictive than those of the Genon theory. We have arrived at the definition of a gene as the pre-image of elementary functional units on the heritable material. Abandoning the identification of function with a functional product, we highlight the logical separation between functions (measured by some experimental protocol) and expression products. Expression of products, as described in “Gene expression as computation”, is understood as computation-like processing cascade that starts with the generation of a working copy of the inheritable genetic information. The understanding of the mechanics of expression (or the corresponding computation) does not require the notion of a gene at all. It is sufficient to consider the processing products and their molecular interactions. Indeed, a sufficiently detailed model of the expression processes is likely to be a good starting point to define function, functional units, and eventually genes.
The precise meaning of the term “gene expression” remains elusive. Logically, it refers to the construction of functional units from their heritable source. Since genes are not synonymous with “products in the expression cascade”, gene expression is not synonymous with the processing of individual transcripts (or other individual processing products). Instead, it must be understood as a composite of the expression program governing the construction of the molecular components of the functional unit, together with additional interactions that are not encapsulated in any expressed molecular product. A simple one-to-one relation between the chemical and logical expression programs exists only in limiting cases, for instance when functional units are identified with polypeptides as in the Genon Theory. In general, it remains to be seen to what extent (logical) gene expression can be modeled in a computational framework analogous to the physical expression of products (in the sense of “Gene expression as computation”). Even if gene expression can be modeled in this way, it is not clear a priori how the relations between the physical and the logical expression program can be described.
A simple, but practically relevant implication of the distinction between expressed products and functionally defined genes as advocated here, is that (at least at present) genes are irrelevant for genome annotation. This statement might be perceived as provocative. Nonetheless, we think there are good arguments to take such a radical step. Genome annotation, after all, is a pragmatic enterprise and hence has to concentrate on information that is readily available or can be generated with reasonable efforts. Therefore it is at least largely limited to the physical objects of the expression cascade and information such as binding sites. This information is about biochemical processes at best and is independent of the higher-level biological interpretation. Given the organization of the transcriptome as a complex structure of overlapping products in both reading directions (The ENCODE Project Consortium 2007; Kapranov et al. 2007), it makes little sense to tie a functional interpretation or a disease relevance directly to a DNA position once the functional product involved has been identified. There are, indeed, an increasing number of examples where the same DNA locus gives rise to different products with different functions (Ikeda et al. 2007; Bender 2008). Of course, if the information arose from a mutation or association study, we can only map it to a DNA region, since we do not know the responsible “gene” or expression product.
Acknowledgments
We thank Brendy Alexander, Gene T. Onic, and Margarita A.T. Thepool for stimulating discussions on the gene concept in September 2007, Claudia Copland for comments and editing assistance, and David Krakauer for suggestions on a preliminary version of this manuscript.
Footnotes
Text in italics quotes from Scherrer and Jost (2007a).
Units, whose function(s) rely on input and/or communication of course need to be provided with this stimulus.
“Space” is used here in the formal mathematical sense as “a set endowed with a certain abstract structure.”
For simplicity of language we speak of the “genomic source” instead of the more general “encoding in the inheritable material”.
Contributor Information
Sonja J. Prohaska, Email: sonja@santafe.edu
Peter F. Stadler, Email: studla@bioinf.uni-leipzig.de
References
- Attiya H, Welsh J (2004) Distributed computing: fundamentals, simulations, and advanced topics. Wiley, New York
- Bender W (2008) MicroRNAs in the Drosophila bithorax complex. Genes Dev 22:14–19 [DOI] [PMC free article] [PubMed]
- Brent MR (2005) Genome annotation past, present, and future: How to define an ORF at each locus. Genome Res 15:1777–1786 [DOI] [PubMed]
- Carninci P (2006) Tagging mammalian transcription complexity. Trends Genetics 22:501–510 [DOI] [PubMed]
- Carrasco CD, Buettner JA, Golden JW (1995) Programed DNA rearrangement of a Cyanobacterial hupL gene in Heterocysts. Proc Natl Acad Sci USA 92:791–795 [DOI] [PMC free article] [PubMed]
- Danos V, Laneve C (2004) Formal molecular biology. Theor Comput Sci 325:69–110 [DOI]
- Danos V, Feret J, Fontana W, Harmer R, Krivine J (2007) Rule-based modelling of cellular signalling. In: Caires L, Vasconcelos VT (eds) CONCUR 2007—concurrency theory, 18th international conference, vol 4703 of Lecture Notes in Computer Science. Springer, Heidelberg, pp 17–41
- El-Sharoud WM, Graumann PL (2007) Cold shock proteins aid coupling of transcription and translation in bacteria. Sci Prog 90:15–27 [DOI] [PMC free article] [PubMed]
- Garnier O, Serrano V, Duharcourt S, Meyer E (2004) RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol 24:7370–7379 [DOI] [PMC free article] [PubMed]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M (2007) What is a gene, post-ENCODE? history and updated definition. Genome Res 17:669–681 [DOI] [PubMed]
- Gowrishankar J, Harinarayanan R (2004) Why is transcription coupled to translation in bacteria? Mol Microbiol 54:598–603 [DOI] [PubMed]
- Ikeda Y, Daughters RS, Ranum LP (2007) Bidirectional expression of the SCA8 expansion mutation: One mutation, two genes. Cerebellum. doi:10.1080/14734220701413781 [DOI] [PubMed]
- Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21:1833–1856 [DOI] [PubMed]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356 [DOI] [PubMed]
- Kapranov P, Cheng J, Dike S, Nix D, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Madhavan G, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488 [DOI] [PubMed]
- Kuttler C, Niehren J (2006) Gene regulation in the Pi Calculus: simulating cooperativity at the lambda switch. In: Transactions on computational systems biology VII, vol 4230 of Lecture Notes in Computer Science. Springer, Heidelberg, pp 24–55
- Kvasnicka V, Pospíchal J (2002) Emergence of modularity in genotype-phenotype mappings. Artif Life 8:295–310 [DOI] [PubMed]
- Lewin B (1994) Genes V. Oxford Univ Press, Oxford
- Listerman I, Sapra AK, Neugebauer KM (2006) Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13:815–822 [DOI] [PubMed]
- Lolle SJ, Victor JL, Young JM, Pruitt RE (2005) Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 434:505–509 [DOI] [PubMed]
- Maciag K, Altschuler SJ, Slack MD, Krogan NJ, Emili A, Greenblatt JF, Maniatis T, Wu LF (2006) Systems-level analyses identify extensive coupling among gene expression machines. Mol Syst Biol 3:0003 [DOI] [PMC free article] [PubMed]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engström PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, Bult CJ, Fletcher CF, Forrest AR, Furuno M, Hill D, Itoh M, Kanamori-Katayama M, Katayama S, Katoh M, Kawashima T, Quackenbush J, Ravasi T, Ring BZ, Shibata K, Sugiura K, Takenaka Y, Teasdale RD, Wells CA, Zhu Y, Kai C, Kawai J, Hume DA, Carninci P, Hayashizaki Y (2006) Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genetics 2:62. doi:10.1371/journal.pgen.0020062 [DOI] [PMC free article] [PubMed]
- Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW (2007) Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci 120:2755–2761 [DOI] [PubMed]
- Pheasant M, Mattick JS (2007) Raising the estimate of functional human sequences. Genome Res 17:1245–1253 [DOI] [PubMed]
- Sambrook J, Russel D (2001) Molecular cloning: a laboratory manual. Cold Spinger Harbor Laboratory Press, Cold Spring Harbor
- Scherrer K, Jost J (2007a) The gene and the genon concept: a conceptual and information-theoretic analysis of genetic storage and expression in the light of modern molecular biology. Theory Biosci 126:65–113 [DOI] [PMC free article] [PubMed]
- Scherrer K, Jost J (2007b) The gene and the genon concept: a functional and information-theoretic analysis. Mol Syst Biol 3:87 [DOI] [PMC free article] [PubMed]
- Schlosser G (2002) Modularity and the units of evolution. Theory Biosci 121:1–80 [DOI]
- Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference—a crash course. Trends Genet 21:339–345 [DOI] [PMC free article] [PubMed]
- Snyder M, Gerstein M (2003) Genomics: defining genes in the genomics era. Science 300:258–260 [DOI] [PubMed]
- Swinburne IA, Meyer CA, Liu XS, Silver PA, Brodsky AS (2006) Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res 16:912–921 [DOI] [PMC free article] [PubMed]
- Tanaka RJ, Okano H, Kimura H (2006) Mathematical description of gene regulatory units. Biophys J 91:1235–1247 [DOI] [PMC free article] [PubMed]
- The ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816 [DOI] [PMC free article] [PubMed]
- Wagner GP, Pavlicev M, Cheverud JM (2007) The road to modularity. Nat Rev Genet 8:921–931 [DOI] [PubMed]
- Willingham AT, Gingeras TR (2006) TUF love for “junk” DNA. Cell 125:1215–1220 [DOI] [PubMed]
- Zufall RA, Robinson T, Katz LA (2005) Evolution of developmentally regulated genome rearrangements in eukaryotes. J Exp Zool Mol Dev Evol 304B:448–455 [DOI] [PubMed]