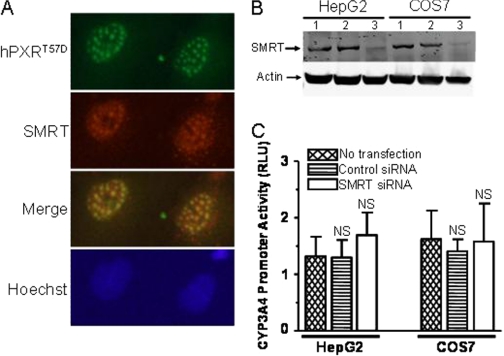
Fig. 5.
Silencing of SMRT cannot rescue the impaired function of hPXRT57D. A, hPXRT57D colocalizes with SMRT to the nucleus at discrete nuclear foci. COS7 cells were transfected with FLAG-hPXRT57D. Twenty-four hours post-transfection, the cells were treated with vehicle 0.1% DMSO or 10 μM rifampicin. The cells were processed for immunocytochemistry 24 h after treatment with DMSO or rifampicin. FLAG-hPXRT57D was probed using rabbit anti-hPXR polyclonal antibody; SMRT was probed using anti-SMRT mouse monoclonal antibody; and nuclear DNA was stained with Hoechst dye. Data shown are from a representative experiment. B, Accell SMARTpool human SMRT siRNA knocks down the protein expression of SMRT. HepG2 and COS7 cells were transfected with 1 μM Accell nontargeting pool control siRNA (lane 2) or Accell SMARTpool human SMRT siRNA (lane 3) in Accell siRNA delivery media. Whole cell lysates were collected 72 h after transfection and subjected to Western blotting analysis using anti-SMRT and anti-actin antibodies. Protein expression for SMRT without transfection was shown in lane 1. Actin protein expression was shown as a loading control. Data shown are from a representative experiment. C, silencing of SMRT cannot rescue the impaired transactivation function of hPXRT57D. HepG2 and COS7 cells were transfected with pcDNA3-FLAG-PXRT57D, pGL4-hRluc, and pGL3-CYP3A4-luc using Lipofectamine. Five hours after transfection with the plasmids, the cells were transfected with 1 μM Accell nontargeting SMARTpool control siRNA or Accell SMARTpool SMRT siRNA in Accell siRNA delivery media for 72 h. Forty-eight hours after transfection with the siRNAs, the cells were treated with vehicle DMSO (0.1%) or 10 μM rifampicin. Firefly and Renilla luciferase activities were measured 24 h after rifampicin treatment. Firefly luciferase activity was first normalized with Renilla luciferase activity before normalizing the data for rifampicin with DMSO. Data are shown as mean values from four independent experiments with bars indicating the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the RLU obtained from the samples transfected with control siRNA or SMRT siRNA with the samples that were not transfected with siRNA. Differences were considered nonsignificant (N.S.) for p > 0.05.