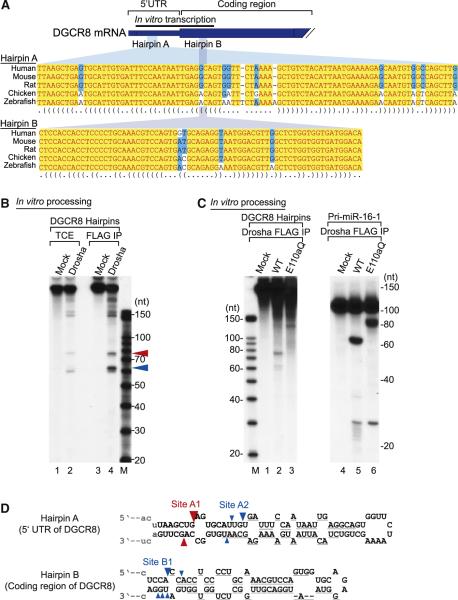
Figure 2. The Hairpins in DGCR8 mRNA Are Cleaved by Drosha In Vitro.
(A) Partial sequences of DGCR8 mRNA of human, mouse, rat, chicken, and zebrafish. There are two hairpin structures, hairpin A in the 5′ UTR and hairpin B in the coding region. A black bar presents a template for in vitro transcription. This figure is adapted from Pedersen et al. (2006).
(B) In vitro processing of the DGCR8 hairpins. Internally labeled hairpins were incubated with total cell extract (TCE) or FLAG-immunopurified (FLAG-IP) Drosha. Cleavage fragments are marked by red or blue triangles.
(C) In vitro processing of the DGCR8 hairpins with a Drosha mutant E110aQ. Pri-miR-16-1 was used as a control.
(D) Cleavage site mapping of the two hairpins. The cleavage sites are indicated with triangles. The color of the triangles shows the origin of the fragments presented in Figure 2B. Size of the triangle reflects the clone frequency such that the frequencies of 99%-75%, 74%-50%, and 49%-25% are shown with large, medium, and small triangles, respectively. The cleavage sites with clone frequencies of under 25% are not presented in this figure. Gray lines under the hairpins represent the binding sites for the probes used for northern blotting.