Fig. 3. PAX2 binds to the hBD1 promoter in vitro.
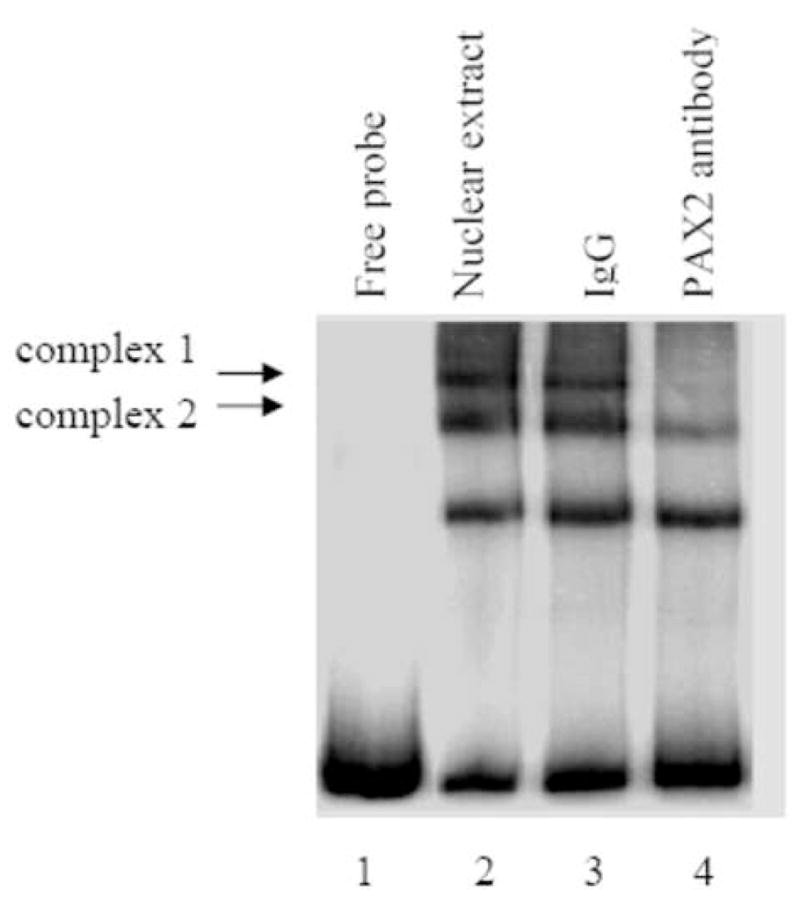
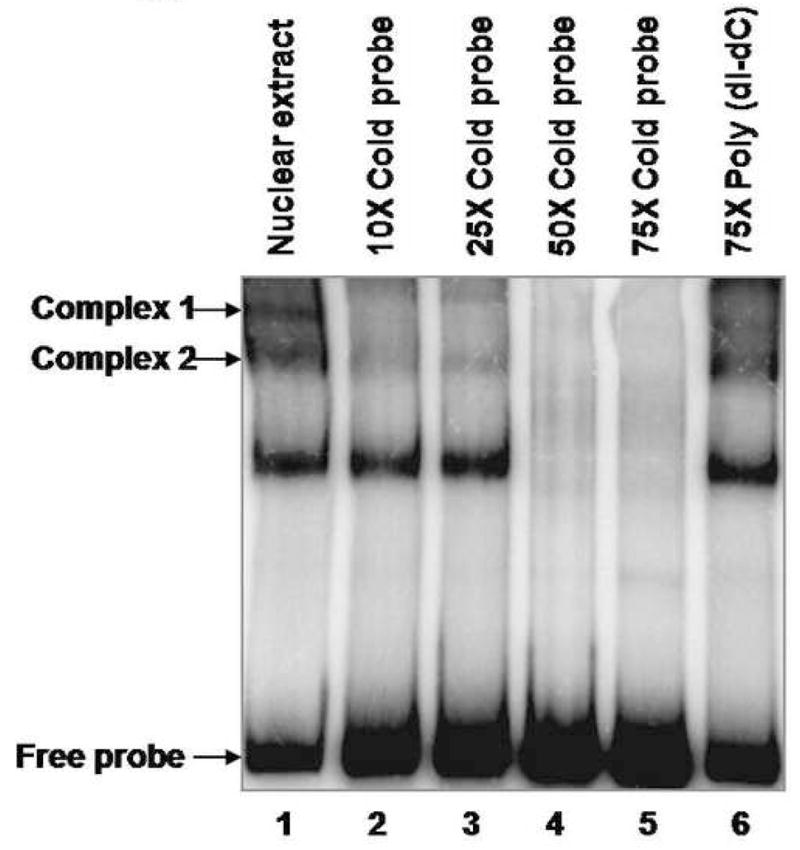

A, Electrophoretic mobility shift assay of the 32P-hBD1 DNA probe without nuclear extract sample (lane 1), or in the presence of 12 μg of nuclear extract from DU145 cells (lane 2). The presence of PAX2 in protein-DNA complexes was tested by adding anti-PAX2 antibody to the reaction mixture (lane 4) which showed decrease in intensity of two complexes. However, incubations of nuclear extract with normal rabbit IgG, no change in the intensities of the complexes was observed. B, Gel mobility shift competition assay was performed to test the specificity of PAX2 binding to hBD1-promoter. Electrophoresis of the 32P-labeled hBD1-DNA with DU145 nuclear extract showed bands of DNA-protein complexes (lane 1). To examine the specificity of DNA binding, nuclear extract was incubated with 32P-hBD1 DNA in the presence of 10X (lane 2), 25X (lane 3), 50X (lane 4) and 75X (lane 5) molar excess of unlabeled competitor homologous hBD1-DNA or 75X molar excess of poly-dI-dC as a negative control (lane 6). C, SiRNA knock-down of PAX2 was performed to determine the effect of PAX2 suppression on protein-DNA complex expression. Electrophoresis of the 32P-labeled hBD1-DNA without nuclear extract (lane 1) or incubated with nuclear extract from DU145 cells treated with scramble siRNA (lane 2) or PAX2 siRNA (lane 3). After incubation in binding buffer, samples were separated on a 5% acrylamide gel which was dried and analyzed by phosphor-imaging.
