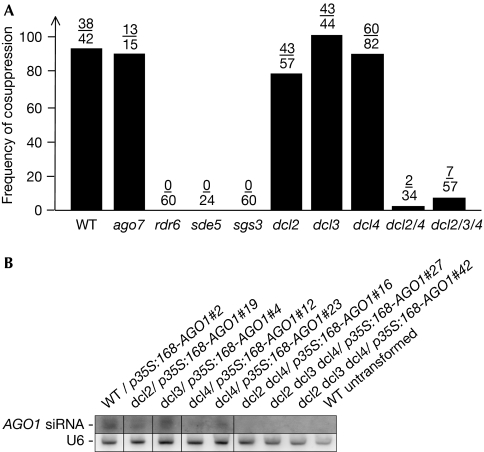
Figure 3.
AGO1 cosuppression requires SGS3, SDE5, RDR6, DCL2 and DCL4. (A) Frequencies of AGO1 cosuppression observed after the transformation of wild-type (WT) plants and the indicated mutants with p35S:168-AGO1. The fraction of plants showing the cosuppression phenotypes of AGO1 is indicated above each bar. (B) RNA gel blot analysis of AGO1 siRNAs in WT untransformed and p35S:168-AGO1-transformed WT and dcl mutant plants showing strong cosuppression of developmental defects. The accumulation of AGO1 siRNA depends on both DCL2 and DCL4. For both dcl4 and dcl2 dcl3 dcl4, two independent transformants are presented. The blot was hybridized for U6 snRNA as a control. AGO, ARGONAUTE; DCL, DICER-LIKE; RDR6, RNA DEPENDENT RNA POLYMERASE 6; SDE5, SILENCING DEFECTIVE 5; SGS3, SUPPRESSOR OF GENE SILENCING 3; siRNA, short interfering RNA; snRNA, small nuclear RNA.