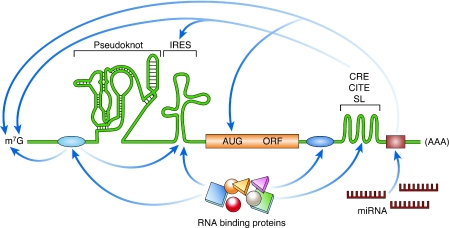
Figure 1.
Schematic illustration of discreet RNA regulatory elements. The translation of an mRNA is regulated by diverse mechanisms that involve both structural and non-structural RNA elements, as well as interactions with RNA-binding proteins. IRESes are found in the 5′ UTR and promote cap-independent translation. Pseudoknots can be located in the 5′ UTR, the 3′ UTR, or the coding region, and their localization influences their effect on translation; for example, initiation, frameshifting and termination. 3′ UTR structural elements such as CREs, CITEs and SLs often function through long-range RNA interactions. The miRNA target sites are located in the 3′ UTR. The canonical mRNA features of the cap (m7G) and poly(A) are targets of several regulatory interactions. The RNA-binding protein-binding sites are shown as blue ovals, and the miRNA-binding site is shown as a brown rectangle. AUG, initiation codon; CITE, cap-independent translational enhancer; CRE, cis-acting replication element; IRES, internal ribosome entry site; m7G, 7-methyl-guanosine; miRNA, microRNA; mRNA, messenger RNA; ORF, open-reading frame; SL, stem loop; UTR, untranslated region.