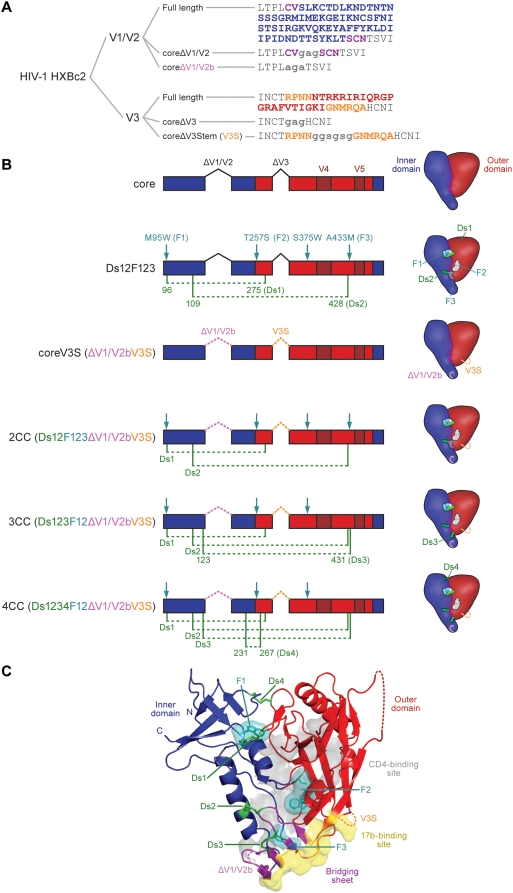
Figure 1. Structural elements of the HXBc2 gp120 core protein and its stabilized derivatives.
A. Amino acid sequences of the V1/V2 loop and the V3 loop as present in full-length gp120, in the previously crystallized core protein and in coreV3S protein. Linker sequences are in lower case. B. Linear diagram of core and coreV3S derivatives showing positions of V1/V2- and V3 loop deletions, cavity-filling mutations (F, teal arrows) and paired disulfide mutations (Ds, green dotted lines). The schematics depict surface models of each corresponding core derivative indicating relative positions of the mutations within inner domain (blue) and outer domain (red). C. Ribbon diagram of HXBc2 core gp120 crystal structure with all stabilizing mutations modeled on it. Indicated are the modifications of V1V2- and V3 loops (labeled dashed lines), the CD4 binding surface (translucent gray), the 17b epitope surface contacts (yellow) and the beta strands comprising the bridging sheet sub-domain (purple). Note the proximity of Ds2 and Ds3 to both the CD4 binding site and the bridging sheet. 17b is the prototypic co-receptor-binding-site-directed antibody, which inhibits gp120-CD4 interaction with co-receptor, and serves as surrogate for co-receptor N-terminal interaction with the gp120 core. Conserved V3 loop elements (not shown) also contribute to gp120-CD4-co-receptor interaction.