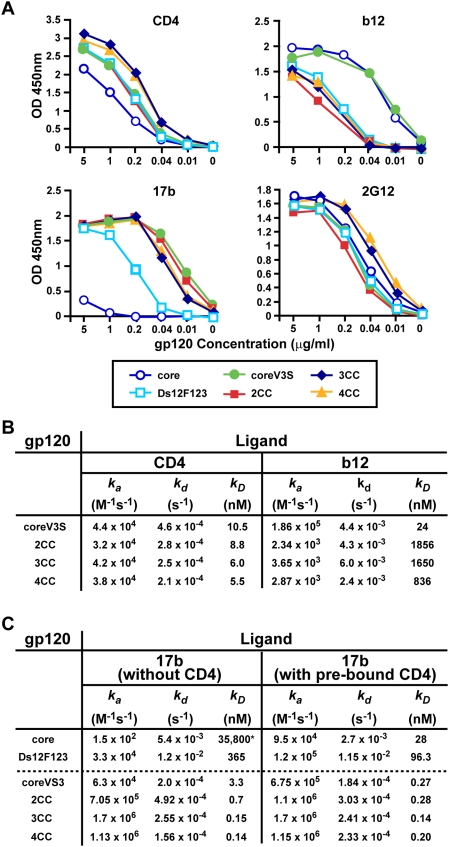
Figure 2. Antigenic analysis of unmodified and structurally stabilized core envelope variants by ELISA and SPR.
A. ELISA plates were coated with ligand (2 µg/ml), reacted with 5-fold serial dilutions of affinity-purified envelope glycoproteins (starting at 5 µg/ml) and detected with 1∶2500 dilution of rabbit immune sera raised against HXBc2 core protein (unmodified). Upper left, binding to soluble human CD4 (4-domain; Progenics); upper right, binding to b12; lower left, binding to 17b; lower right, binding to 2G12. Margins of error from duplicate wells were negligible in all cases. B and C. Binding rate constants for interactions of stabilized gp120 core proteins with sCD4, b12 and 17b. Approximately 500 RU each of 17b, sCD4 and b12 were coated on CM5 chip. Two-fold serial dilutions of each gp120 protein were allowed to bind to the surfaces for 3 min followed by dissociation for 5 min. The kinetic constants were obtained by fitting the curves to 1∶1 Langmuir binding model. B. Kinetics of CD4 and b12 interactions. C. Kinetics of 17b binding to gp120 variants without or with pre-exposure to 10-fold molar excess of sCD4. *Data obtained from very low binding interaction (maximum RU of 8.5).