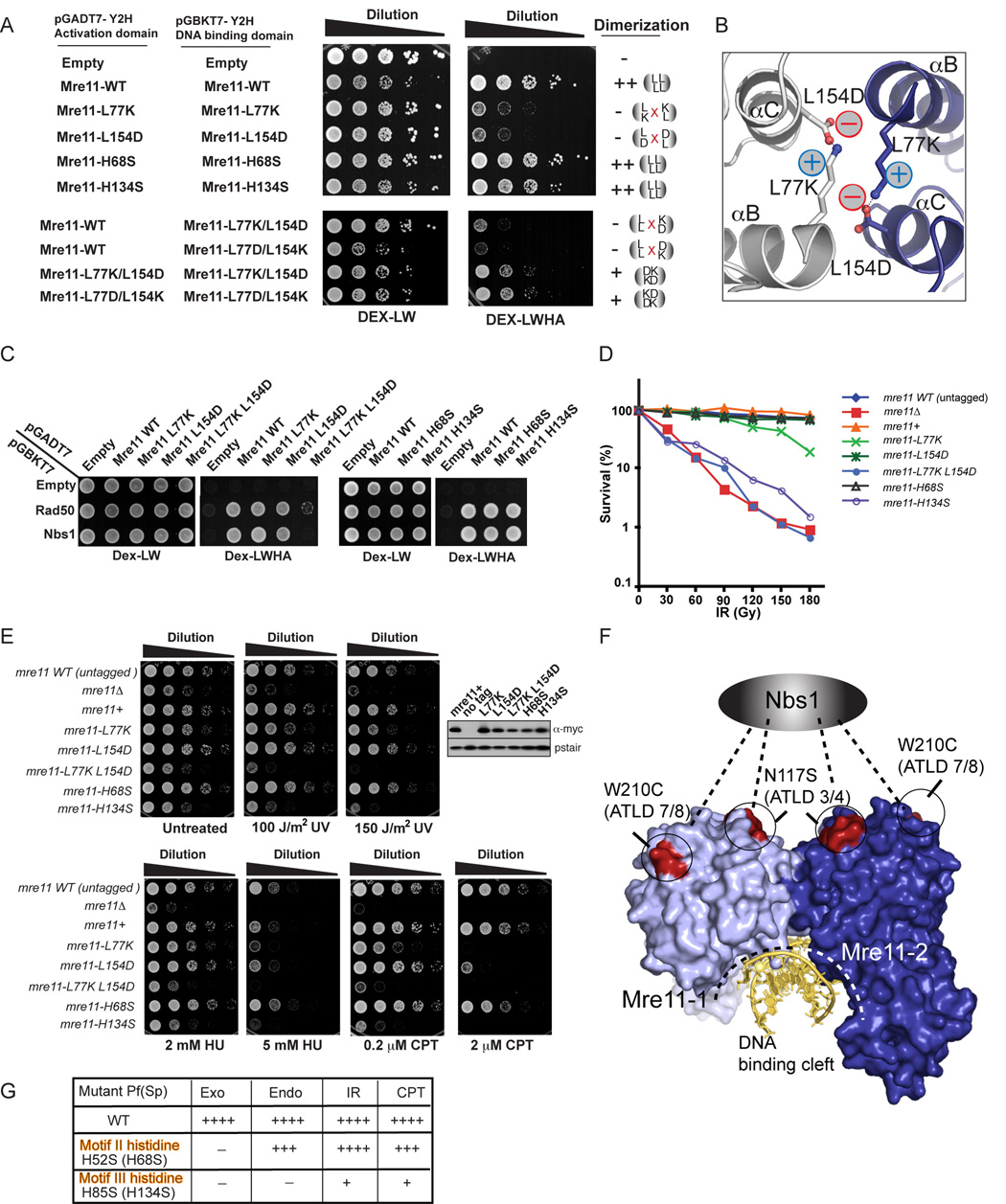
Figure 5. Dimerization and Nuclease Activities are Required for Mre11 complex DNA Repair Functions in S. pombe.
(A) Mre11 self-interaction by two hybrid assays. Growth on DEX-LWHA indicates a positive two-hybrid interaction.
(B) Energy minimized model for a designed salt-bridged, reconstituted Mre11-Mre11 dimeric interface.
(C) Mre11 variant interactions with Rad50 and Nbs1 by two-hybrid analyses. Growth on Dex-LWHA indicates a positive two-hybrid interaction.
(D) Mre11 dimerization and endonuclease disruption variants are IR sensitive.
(E) CPT, HU, and UV, and show that disruption (Mre11-L77K) or distortion (Mre11-L77K L154D) of the Mre11 dimer interface or catalytic histidine mutant H134S results in clastogen sensitivity. Five-fold serial dilutions of cells on YES plates were photographed after 2–3 days growth at 30°C. Expression levels of myc-tagged wild type and mutant Mre11 proteins were determined by immuno-blotting.
(F) ATLD missense mutations W210C and N117S, which impair Nbs1 binding, cluster to a single Mre11 dimer surface opposite the DNA binding cleft.
(G) Endonuclease activity is required for efficient DSB repair in fission yeast. Mre11 catalytic activity for mutant P. furiosus proteins is compared to genotoxin resistance levels (++++ for wild-type to + for highly sensitive but above mre11−) in fission yeast for Motif II and Motif III histidine mutant yeast strains (bracketed mutations).