Abstract
Objectives. To report on a novel IIF pattern specifically associated with antibodies to DNA topo I.
Methods. A novel compound IF pattern, designated Scl-70 pattern, was characterized in routine ANA-HEp-2 IIF screening. Within the last 3 years, all serum samples presenting the Scl-70 pattern at the ANA-HEp2 IIF screening were tested for anti-topo I reactivity. Conversely, 16 serum samples with known anti-topo I reactivity and affinity-purified anti-topo I antibody preparations were tested for the Scl-70 pattern.
Results. The Scl-70 pattern comprised the staining of five cellular regions: nucleus, nucleolus and cytoplasm in interphase cells; nucleolar organizing region (NOR) and chromosomes in mitotic cells. All 81 serum samples selected as Scl-70 pattern reacted with topo I. All 16 anti-topo I samples and antibody preparations reproduced the Scl-70 pattern. This compound IF pattern was consistently observed in different commercial HEp-2 cell slides and in home-made slides with HEp-2 cells and human fibroblasts fixed with alternative protocols. Double IIF experiments demonstrated the co-localization of topo I and human upstream binding factor at the NOR.
Conclusions. The Scl-70 pattern belongs to the group of compound IF patterns that hold strong association with the respective autoantibody specificities, such as that observed with centromere protein F (CENP-F) and nuclear mitotic apparatus-1 (NuMA-1) protein. The identification of the Scl-70 pattern at routine ANA-HEp-2 IIF screening may lead to implementation of specific tests for the identification of anti-topo I antibodies. In addition, the Scl-70 pattern outlines cellular domains other than those previously reported for topo I, which is of interest for further understanding the roles of this enzyme in cell biology.
Keywords: Anti-nuclear antibodies, DNA topoisomerase I, Scl-70, Autoantibodies, Systemic sclerosis
Introduction
The standard method of screening for ANAs has been the IIF assay on HEp-2 cells (ANA-HEp-2 IIF) [1]. In addition to its usefulness as a screening tool, the ANA-HEp-2 IIF assay is also able to provide hints for the autoantibody specificity in a given sample. HEp-2 cell line (American Type Culture Collection CCL23, Rockville, MD, USA) is derived from human epithelial carcinoma and grows in monolayers of widely spread cells that nicely display a well-organized array of cell domains. Each cell domain is enriched with a distinct set of antigens. Therefore, the IF pattern depicted with a given serum is a function of the topographic distribution of the antigens recognized by the antibodies present in that serum. In fact, it has been broadly recognized that some IF patterns are associated with certain autoantibodies. For example, the nuclear coarse speckled pattern is strongly associated with anti-Smith (anti-Sm) and anti-snRNP antibodies [2,3], and the nuclear homogeneous pattern is associated with antibodies to native DNA, nucleosome and histones [4].
Since cell domains contain several molecular components that may be target of autoantibodies, a given IF pattern is usually associated with more than one autoantibody specificity. For example, the IF pattern associated with the Golgi complex may be elicited by autoantibodies to several of the Golgi constituents, such as giantin/macrogolgin/GCP372, golgin-245/p230, GMAP-210, golgin-160/GCP170, golgin-95/GM130 and golgin-97 [5–10]. The IF pattern associated with the nucleolus may be elicited by autoantibodies to several nucleolar constituents, such as Th/To, fibrillarin, NOR90 (nucleolar organizing region-90), RNA polymerase I and nucleolin, involved in various stages of ribosomal biosynthesis and processing [11–13]. Therefore, the ANA-HEp-2 IF pattern may provide hints on the autoantibody specificity, but in general additional tests are required for the definite autoantibody identification.
However, there are some exceptions in which the IF pattern is strongly associated with a given autoantibody specificity. In fact, some autoantigens are expressed in more than one cell domain and some of these may undergo repositioning along the cell cycle. The resulting compound topographic distribution may be such that it is not shared by other autoantigens located in any of the isolated cellular domains that make up the compound pattern. Compound IF patterns may present such a strong association with the target autoantigens that they may be interpreted as virtually specific for the respective autoantibodies. One example is the characteristic pattern displayed by antibodies to nuclear mitotic apparatus (NuMA-1) protein, which is represented by a compact fine speckled staining of the nucleus of interphase cells and a peculiar staining of the mitotic poles and spindle fibres in metaphase cells [14]. Samples staining the mitotic poles without the concomitant fine speckled staining of interphase nucleus are not associated with NuMA-1 antibodies and none of the samples with anti-NuMA-1 antibodies failed to present the characteristic NuMA-1 compound IF pattern. Another example is the compound pattern associated with anti-centromere protein F (anti-CENP-F) antibodies [15]. This highly pleomorphic pattern is characterized by heterogeneous fine speckled staining of the interphase nucleus, centromeric staining restricted to mitotic cells and a characteristic staining of the mid-body and intercellular bridge. Virtually all samples presenting this specific pattern are reactive to CENP-F [15].
Autoantibodies reacting with DNA topo I are serological markers of SSc. They are observed in up to 30% of the unselected SSc patients and are associated with diffuse skin involvement and more aggressive forms of the disease [16]. Anti-DNA topo I antibodies have been traditionally associated with an IF pattern on HEp-2 cells characterized by nuclear and nucleolar fine speckled staining of interphase cells. In addition, mitotic cells display a fine speckled staining of the metaphase plate [17,18]. Although usually observed with sera containing anti-DNA topo I antibodies, this pattern is not considered specific for these autoantibodies. In our routine ANA-HEp-2 IIF analysis, we have observed additional features in the IF pattern associated with anti-DNA topo I antibodies. This compound IF pattern, herein designated as Scl-70 pattern [original designation for the autoantigen (DNA topo I) recognized by some SSc sera], comprises the staining of five cellular domains along the cell cycle. The objective of this report is to characterize the compound Scl-70 pattern and to demonstrate its strong association with anti-DNA topo I antibodies.
Materials and methods
Within the last 3 years, all serum samples presenting the Scl-70 pattern (see Results section for details) at the routine ANA–HEp2 screening were investigated for the presence of anti-DNA topo I antibodies. These comprised a total of 81 Scl-70 pattern samples. In addition, serum samples with known reactivity to DNA topo I and purified anti-DNA topo I antibody preparations were tested in the ANA-HEp-2 IIF assay for the presence of the Scl-70 pattern. These comprised 16 samples of patients with SSc defined according to the standard criteria [19], the centers for disease control (CDC) reference human anti-DNA topo I serum (No. IS—2135 lot No. 84-0027, ANA Reference Human Serum No. 9 pos. anti-Scl 70), commercial positive anti-DNA topo I standard control (Bio-Rad Laboratories, Hercules, CA, USA), Sepharose affinity-purified anti-DNA topo I antibody preparation (kindly donated by Dr R. W. Burlingame, Inova Diagnostics, San Diego, CA, USA) and western blot-based affinity-purified anti-DNA topo I antibody preparation. Affinity purification of anti-DNA topo I antibodies from SDS–PAGE nitrocellulose membranes was performed as previously described [20] using a high titre SSc serum sample with anti-DNA topo I reactivity determined by double ID and western blot. Informed consent was obtained from patients with SSc, who donated serum samples for the experiments. The study was approved by the ethics committee at Fleury Research and Development Institute.
IIF was performed at the dilution of 1 : 80 according to the standard protocol [1] on a variety of cell substrates, including commercial HEp-2 cell slides from six different suppliers [Hemagen Diagnostics, Bion Enterprise, Bio-Rad Laboratories (Columbia, MD, USA), Inova Diagnostics, The Binding Site (Birmingham, UK) and IMMCO Diagnostics (Buffalo, NY, USA)], home-made HEp-2 cell slides and home-made human dermal fibroblasts slides. For the home-made slides, HEp-2 cells (ATCC, American Type Culture Collection CCL23) were grown to subconfluency and transferred to appropriate 12-well glass slides. After an additional 24-h period at DMEM (Gibco Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (Gibco Invitrogen Life Technologies) at 5% CO2 and 37°C, cells were fixed in methanol at −20°C for 5 min and acetone at −20°C for 2 min. Alternatively, cells were fixed in 4% paraformaldehyde for 30 min at room temperature, washed three times in 0.15 M phosphate buffered saline (PBS) of pH 7.2, incubated in 50 mM ammonium chloride for 15 min, permeabilized in acetone at −20°C for 2 min and kept in PBS at 4°C until use. Primary human fibroblast culture was obtained by explant from forearm biopsy of normal volunteers and grown on round glass cover slips to subconfluency. Fixation in paraformaldehyde was performed as above. All slides were analysed by three independent observers in an Olympus (Center Valley, PA, USA) B ×50 fluorescence microscope at × 400 magnification. Images captured with a QColor 3 camera (Olympus) were analysed with the Image-J and Image-Pro Plus 5.1 software (Media Cybernetics, Bethesda, MD, USA).
For double IIF, commercial HEp-2 slides were incubated with anti-human upstream binding factor (anti-hUBF) murine monoclonal IgG (F-9 Sc-13 125, lot L1703, Santa Cruz Biotechnology, Santa Cruz, CA, USA) in the ratio 1 : 100 for 1 h at 37°C, washed in PBS and incubated with different anti-DNA topo I human serum samples diluted 1 : 80 for 1 h at 37°C. Alternatively, cells were incubated with affinity-purified anti-DNA topo I antibody preparations diluted 1 : 5 (western blot purification) and 1 : 100 (protein A–Sepharose purification) in PBS. After a further washing step as before, cells were incubated with anti-murine IgG (Alexa Fluor 568 mouse, A11031 L84E2-1, Invitrogen, Molecular Probes) and anti-human IgG in 1 : 100 ratio (FITC, Bion Enterprise, Des Plaines, IL, USA, CCP-0269) for 1 h at 37°C. After a final washing step, slides were mounted with buffered glycerin and 4′,6-diamidino-2-phenylindole dihydrochloride II (DAPI II) (125 ng/ml, Abbott Molecular, # 30-804 931) and analysed in a scanning confocal fluorescence system (Bio-Rad Laboratories MRC1024 UV) attached to a Zeiss Axiovert 100 microscope, using either a 40× 1.2 NA water immersion or a 100× 1.4 oil immersion PlanApochromatic objective.
Antibodies to DNA topo I were determined by standard double ID assay and western blot. The ID assay was performed as previously reported [21] with rabbit thymus extract (Pel Freez Biologicals, Rogers, AR, USA) and secondary standard anti-DNA topo I serum previously established from the sample provided by the Centers for Disease Control (No. IS-2135 lot No. 84-0027; CDC, Atlanta, GA, USA). Western blot was performed with HEp-2 whole cell extract separated in 10% SDS–PAGE according to the previously established method [22]. Briefly, serum samples were tested at 1 : 50 dilution in 5% skim milk 0.05% Tween-20 in PBS (MT-PBS) for 1 h at room temperature under shaking. Alternatively, strips were incubated with western blot-based affinity-purified anti-DNA topo I antibody preparation diluted to the ratio 1 : 5 in MT-PBS. After two 15-min washing steps in 0.05% Tween–PBS, nitrocellulose strips were incubated with horseradish peroxidase-labelled anti-human IgG (Bio-Rad Laboratories) diluted to the ratio 1 : 1500 in MT-PBS for 1 h at room temperature in the dark under shaking. After washing with 0.05% Tween–PBS for 15 min and with PBS for 15 min, strips were incubated with chromogenic solution (6 mg 4-chloro-1-naphthol in 2 ml methanol, 10 ml PBS and 20 µl 30% H2O2). The reaction was stopped with water.
Results
Definition of the Scl-70 IF pattern
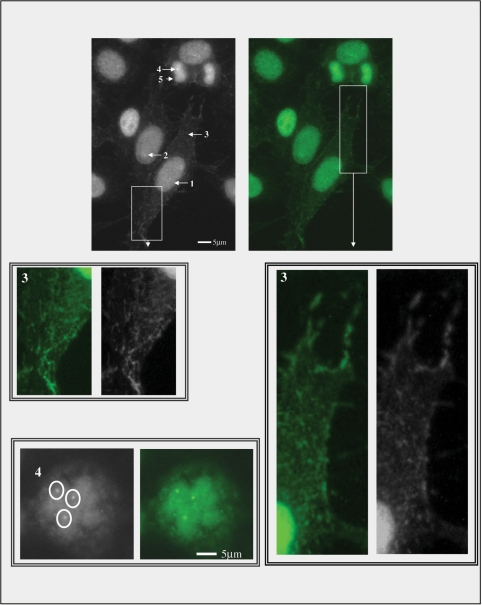
The IF pattern designated Scl-70 pattern comprises the staining of five cell domains: nucleus, nucleolus, cytoplasm in interphase cells, NOR and chromosome metaphase plate in mitotic cells (Fig. 1). The interphase nucleus is stained as a compact fine speckled pattern and there is a faint and inconsistent enhancement of the staining at the nucleolus. The cytoplasm depicts a weakly stained network from the perinuclear area towards the plasma membrane. The chromosome mass in mitotic cells is consistently stained. In addition, there is an enhanced staining at discrete spots at the metaphase plate, resembling the NOR domains (Fig. 1). This compound IF pattern was consistently observed in all commercial HEp-2 cell slides, in home-made HEp-2 cells fixed with the methanol/acetone and with the paraformaldehyde protocols and in human fibroblasts (data not shown). Double IIF experiments analysed by confocal microscopy demonstrated co-localization of DNA topo I reactivity and human upstream binding factor at the NOR at metaphase, anaphase and telophase (Fig. 2).
Fig. 1.
The compound IF pattern Scl-70 comprises the staining of five cell domains: nucleus (1), nucleolus (2), cytoplasm (3) in interphase cells, NOR (4) and chromosome metaphase plate (5) in mitotic cells. The boxes in white outline blow-up details of the delicate cytoplasm staining. The interphase nucleus is stained as a strong compact fine speckled pattern; the nucleolus is weakly and variably stained; the cytoplasm depicts a weakly stained dot/reticular network from the perinuclear area to the plasma membrane; in mitotic cells the chromosome mass is strongly stained similarly to the interphase nucleus and there is an enhanced staining at discrete spots at the metaphase plate, resembling the NOR domains (inset). IIF on HEp-2 cells (Bion, Des Plaines, IL, USA) with human monospecific anti-Scl-70 serum diluted 1 : 80. Scale bars = 5 µm.
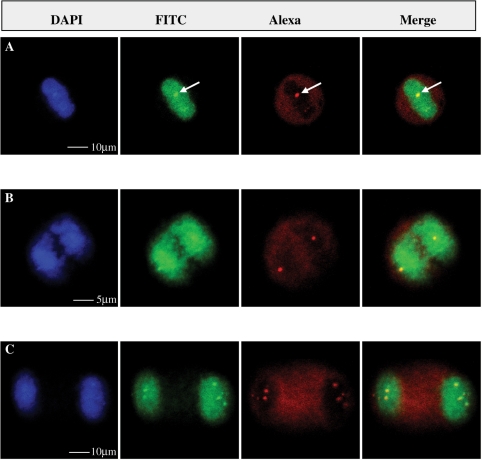
Fig. 2.
Complete co-localization of the domains stained by human anti-DNA topo I serum and NORs in cells at metaphase (A), anaphase (B) and telophase (C). From the left to the right, pictures depict the staining obtained with DAPI, human anti-Scl-70 serum/FITC conjugate, mouse anti-UBF monoclonal antibody/Alexa Fluor 568 conjugate and merging of the latter two images, respectively. Single optical sections acquired by confocal microscopy of IIF on HEp-2 cells (Bion, Park Ridge, IL, USA) with human monospecific anti-Scl-70 serum diluted to 1 : 80 ratio, mouse monoclonal anti-UBF antibody in the ratio 1: 100 and DAPI II (125 ng/ml Abbott Molecular, Des Plaines, IL, USA, 30-804 931). Magnification bars = 5 and 10 µm.
Association of the Scl-70 IF pattern and anti-DNA topo I antibodies
All the 81 serum samples selected in the routine ANA-HEp-2 screening due to the presence of the Scl-70 pattern were confirmed to present anti-DNA topo I antibodies by double ID and by western blot (Fig. 3). Of interest, none of the samples presented the 88–90 kDa doublet characteristic of anti-NOR-90 antibodies. Four samples had the precipitation line on the ID assay formed only after 72 h on diffusion but had indistinguishable characteristics at the ANA-HEp-2 IIF and western blot assays. Conversely, the Scl-70 IF pattern was consistently observed in all 16 SSc serum samples with confirmed anti-DNA topo I antibodies. In addition, the ANA-HEp-2 Scl-70 pattern was also observed with the CDC reference human anti-DNA topo I serum, with the commercial anti-DNA topo I standard serum and with the affinity-purified anti-DNA topo I antibody preparations (Fig. 4). As a control, affinity-purified antibodies from irrelevant areas of the western blot membrane yielded no relevant staining in the HEp-2 IIF assay (data not shown).
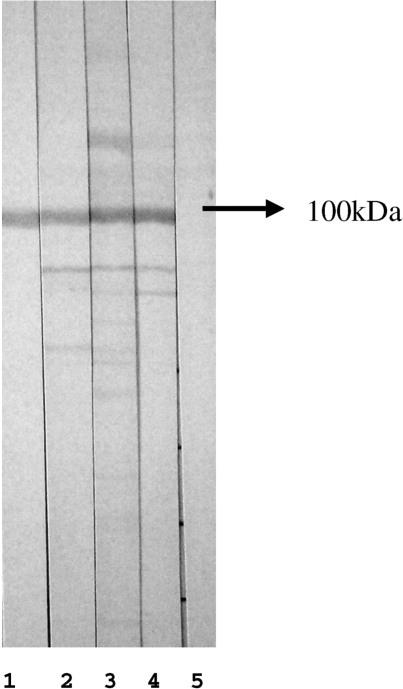
Fig. 3.
Western blot with whole HEp-2 cell extract separated in 10% SDS–PAGE and probed with sera diluted in the ratio 1 : 50. Lane 1: western blot-based affinity-purified antibodies reacting with DNA topo I; lanes 2–4: representative sera depicting the Scl-70 pattern on IIF in HEp-2 cells; lane 5: negative control. Arrow indicates proteins migrating with mobility compatible with 100 kDa.
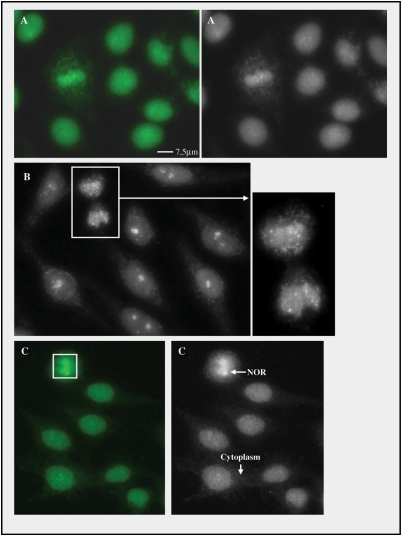
Fig. 4.
The compound IF Scl-70 pattern is reproduced by affinity-purified antibodies to DNA topo I. IIF on HEp-2 cells (Bion) with human monospecific anti-DNA topo I serum diluted to the ratio 1 : 80 (A), western blot-based affinity-purified anti-DNA topo I antibody preparation diluted 1 : 5 (B and inset) and protein A–Sepharose-based affinity-purified anti-DNA topo I antibody preparation diluted 1 : 100 (C). Scale bars = 7.5 µm.
Discussion
In the present study, we originally demonstrated that anti-DNA topo I antibodies present a compound IF pattern encompassing the nucleus, nucleolus, NOR, metaphase chromosome plate and the cytoplasm. This compound pattern was consistently observed in different cellular substrates and with different fixation protocols. The Scl-70 pattern proved to be specific for anti-DNA topo I antibodies, since 81 sequentially selected Scl-70-pattern serum samples were shown to be reactive with DNA topo I in double ID and western blot. In addition, all tested SSc serum samples with known reactivity to DNA topo I presented the ANA-HEp-2 Scl-70 pattern. Finally, affinity-purified anti-DNA topo I antibody preparations could reproduce the Scl-70 pattern.
The recognition of such specific compound IF pattern has important implications from the clinical point of view. The Scl-70 pattern herein characterized belongs to the group of compound IF patterns that hold strong autoantibody specificity association, such as those observed with CENP-F and NuMA-1 autoantibodies [14,15]. The identification of such specific compound patterns in a given sample at the routine ANA-HEp-2 IIF screening leads to the implementation of specific tests for the definitive identification of the respective autoantibody. The HEp-2 cell IF pattern traditionally associated with anti-DNA topo I antibodies refers solely to a fine speckled staining of the nucleus, nucleolus and metaphase plate, which is not exclusively associated with this autoantibody specificity. The Scl-70 pattern herein reported has two additional features (the cytoplasm dot/reticular in interphase cells and the NOR at mitotic cells) that contribute to its specific association with anti-DNA topo I antibodies. The cytoplasmic component of the Scl-70 pattern has never been previously reported and the presence of DNA topo I at the NOR has been previously reported in PtK2 cells but not in HEp-2 cells [23].
The characterization of the compound Scl-70 pattern is also relevant to further understanding of the cell biology of DNA topo I since it indicates the putative cellular domains where the enzyme activity might be involved. DNA topoisomerases are required for relaxation of the DNA helix as it uncoils for DNA replication and RNA transcription. The relaxation process is obtained through DNA cleavage in which a transient phosphodiester bond is formed between a tyrosine residue in the protein and one of the ends of the broken strand. DNA conformation can thus be modified as long as the covalent intermediate persists, and the enzyme is released as the ruptured DNA strand is re-connected. Type I topoisomerases cleave only one DNA strand and type II topoisomerases cleave both DNA strands [24]. Human topo I is a 91 kDa protein of 765 amino acids localized to the nucleus due to the existence of a functional nuclear localization signal [nuclear localization signal (NLS-II), amino acids 150–6] at the N-terminal domain. In addition, there is a second nuclear localization signal (NLS-IV, amino acids 157–99) that is important for nucleolar localization [25].
Chromosome condensation during mitosis involves the generation of right-handed solenoidal supercoils and this implies that topo I would be required at this stage of the cell cycle to relax compensatory negative supercoils [26]. Double mutant yeasts lacking the trf4 and top1 genes are defective in establishing chromosome condensation, spindle elongation and nuclear segregation [27]. These observations may justify the persistence of topo I attached to chromatin during mitosis rendering the characteristic fine speckled staining of the metaphase plate. The enhancement of topo I staining at the NOR in human HEp-2 cell line expands previous similar finding in rat kangaroo PtK2 cells [23]. This morphological finding is supported by previous demonstration of strong binding of DNA topo I to ribosomal DNA (rDNA) in virus-transformed chicken MSB-1 T-cell line [28]. The association of DNA topo I to rDNA may be related to two factors. First, there is some evidence that topo I plays an important role in maintaining the integrity of the rDNA locus [29]. Secondly, RNA polymerases are extremely powerful motors that can probably overcome substantial frictional resistance while translocating along its DNA template in the absence of DNA topoisomerase activity [30]. The cluster organization of rDNA genes might contribute to a paramount DNA helix tension at the rDNA gene loci. Therefore, the enrichment of topo I at the NOR in mitotic cells appears to be appropriate to match the requirement for immediate rRNA transcription right after mitosis [31].
The cytoplasmic component of the Scl-70 pattern has never been reported previously. In fact, such a finding is surprising since DNA topo I is not expected to play any role at cytoplasmic domains. The considerably weaker intensity of the cytoplasmic staining in comparison with the nuclear staining suggests that the observed staining might be due to cross-reactivity of anti-DNA topo I autoantobodies with some cytoplasmic protein. In fact, mitochondria have a specific topoisomerase species [32], and we speculate that this protein might be recognized by anti-DNA topo I autoantibodies. The human mitochondrial topo I gene (TOP1mt) is located at 8q24.3 and consists of 14 exons. The last 13 exons have the exact same size and intron frame as the nuclear topo I gene. The high degree of homology between TOP1mt and nuclear top1 is striking at the amino acid and nucleotide levels. In contrast to the nuclear topo I, mitochondrial topo I lacks an amino acid sequence corresponding to a nuclear localization signal. Its N-terminal sequence has positively charged residues that could fold in a positively charged amphiphilic helix suggestive of a mitochondrial targeting signal. Due to the high amino acid homology of nuclear and mitochondrial topoisomerase species, it is conceivable that the weak dot/reticular cytoplasmic staining observed in the Scl-70 pattern is due to cross-reactivity. This point is currently under investigation in our laboratory.
In conclusion, we have originally reported on a novel compound ANA-HEp-2 IF pattern, the Scl-70 pattern, which is specifically associated with anti-DNA topo I autoantibodies. The recognition of the Scl-70 pattern may be useful in providing relevant information in ANA-HEp-2 IIF screening in the clinical laboratory setting and may contribute to further elucidating the role of this important nuclear enzyme in cell biology.

Acknowledgements
The authors would like to thank Prof. Paulo G. Leser, MD, for helpful comments; Silvia Helena Rodrigues and Silvia Helena Barbosa for selecting and organizing serum samples; and Walter Kindro Andreoli for helping with the preparation of photographic panels. The authors are also grateful to Edward K. L. Chan for expert review and suggestions.
Funding: This study was partially funded by grant 07/50 324-6 from FAPESP (The State of Sao Paulo Research Foundation). Funding to pay the Open Access publication charges for this article was provided by Fleury Medicine and Health.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Tan EM, Feltkamp TE, Smolen JS, et al. Range of antinuclear antibodies in “healthy” individuals. Arthritis Rheum. 1997;40:1601–11. doi: 10.1002/art.1780400909. [DOI] [PubMed] [Google Scholar]
- 2.Rokeach LA, Hoch SO. B-cell epitopes of Sm autoantigens. Mol Biol Rep. 1992;16:165–74. doi: 10.1007/BF00464704. [DOI] [PubMed] [Google Scholar]
- 3.Migliorini P, Baldini C, Rocchi V, Bombardieri S. Anti-Sm and anti-RNP antibodies. Autoimmunity. 2005;38:47–54. doi: 10.1080/08916930400022715. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez C, Garcia-Berrocal B, Herraez O, Navajo JA, Gonzalez-Buitrago JM. Anti-nucleosome, anti-chromatin, anti-dsDNA and anti-histone antibody reactivity in systemic lupus erythematosus. Clin Chem Lab Med. 2004;42:266–72. doi: 10.1515/CCLM.2004.049. [DOI] [PubMed] [Google Scholar]
- 5.Fritzler MJ, Hamel JC, Ochs RL, Chan EK. Molecular characterization of two human autoantigens: unique cDNAs encoding 95- and 160-kD proteins of a putative family in the Golgi complex. J Exp Med. 1993;178:49–62. doi: 10.1084/jem.178.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seelig HP, Schranz P, Schroter H, Wiemann C, Renz M. Macrogolgin—a new 376 kD Golgi complex outer membrane protein as target of antibodies in patients with rheumatic diseases and HIV infections. J Autoimmun. 1994;7:67–91. doi: 10.1006/jaut.1994.1006. [DOI] [PubMed] [Google Scholar]
- 7.Fritzler MJ, Lung CC, Hamel JC, Griffith KJ, Chan EK. Molecular characterization of golgin-245, a novel Golgi complex protein containing a granin signature. J Biol Chem. 1995;270:31262–8. doi: 10.1074/jbc.270.52.31262. [DOI] [PubMed] [Google Scholar]
- 8.Griffith KJ, Chan EK, Lung CC, et al. Molecular cloning of a novel 97-kd Golgi complex autoantigen associated with Sjogren's syndrome. Arthritis Rheum. 1997;40:1693–702. doi: 10.1002/art.1780400920. [DOI] [PubMed] [Google Scholar]
- 9.Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM. GMAP-210, a cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol. 1999;145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nozawa K, Fritzler MJ, Chan EK. Unique and shared features of Golgi complex autoantigens. Autoimmun Rev. 2005;4:35–41. doi: 10.1016/j.autrev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176:117–28. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- 12.Chan EK, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med. 1991;174:1239–44. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S, Alvi A, Holding S, et al. The clinical significance of antinucleolar antibodies. J Clin Pathol. 2008;61:283–6. doi: 10.1136/jcp.2007.049692. [DOI] [PubMed] [Google Scholar]
- 14.Andrade LE, Chan EK, Peebles CL, Tan EM. Two major autoantigen-antibody systems of the mitotic spindle apparatus. Arthritis Rheum. 1996;39:1643–53. doi: 10.1002/art.1780391006. [DOI] [PubMed] [Google Scholar]
- 15.Casiano CA, Humbel RL, Peebles C, Covini G, Tan EM. Autoimmunity to the cell cycle-dependent centromere protein p330d/CENP-F in disorders associated with cell proliferation. J Autoimmun. 1995;8:575–86. doi: 10.1016/0896-8411(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 16.Steen VD, Powell DL, Medsger TA., Jr Clinical correlations and prognosis based on serum autoantibodies in patients with systemic sclerosis. Arthritis Rheum. 1988;31:196–203. doi: 10.1002/art.1780310207. [DOI] [PubMed] [Google Scholar]
- 17.Douvas AS, Achten M, Tan EM. Identification of a nuclear protein (Scl-70) as a unique target of human antinuclear antibodies in scleroderma. J Biol Chem. 1979;254:10514–22. [PubMed] [Google Scholar]
- 18.Jarzabek-Chorzelska M, Blaszczyk M, Jablonska S, Chorzelski T, Kumar V, Beutner EH. Scl 70 antibody–a specific marker of systemic sclerosis. Br J Dermatol. 1986;115:393–401. doi: 10.1111/j.1365-2133.1986.tb06233.x. [DOI] [PubMed] [Google Scholar]
- 19.Subcommittee for Scleroderma Criteria of the American Rheumatism Association Diagnostic & Therapeutic Criteria Committee. Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–90. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 20.Smith DE, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99(1 Pt 1):20–8. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouchterlony O. Antigen-antibody reactions in gels. Acta Pathol Microbiol Scand. 1949;26:507–15. doi: 10.1111/j.1699-0463.1949.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 22.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–19. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guldner HH, Szostecki C, Vosberg HP, Lakomek HJ, Penner E, Bautz FA. Scl 70 autoantibodies from scleroderma patients recognize a 95 kDa protein identified as DNA topoisomerase I. Chromosoma. 1986;94:132–8. doi: 10.1007/BF00286991. [DOI] [PubMed] [Google Scholar]
- 24.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 25.Mo YY, Wang P, Beck WT. Functional expression of human DNA topoisomerase I and its subcellular localization in HeLa cells. Exp Cell Res. 2000;256:480–90. doi: 10.1006/excr.2000.4864. [DOI] [PubMed] [Google Scholar]
- 26.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–33. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 27.Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–76. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- 28.Muller MT, Pfund WP, Mehta VB, Trask DK. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–43. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christman MF, Dietrich FS, Levin NA, Sadoff BU, Fink GR. The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7637–41. doi: 10.1073/pnas.90.16.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 31.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Barcelo JM, Lee B, et al. Human mitochondrial topoisomerase I. Proc Natl Acad Sci USA. 2001;98:10608–13. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]