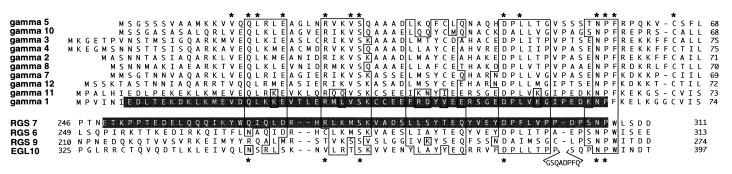
Figure 3.
Structural homology between Gγ subunits and RGS proteins. Alignment of full-length sequences of Gγ subunits and the indicated portions of RGS proteins. Asterisks above the Gγ sequences designate the residues that are identical throughout the entire Gγ class and the residues found at corresponding positions in at least one RGS protein. Boxed are the regions of homology based on the nature of amino acids, i.e., basic (K, R) acidic (E, D), hydrophobic (L, I, V, F, W, M), polar (S, T), and amides (Q, N). EGL-10 has an 8-residue insert shown below its sequence. The sequence highlighted in RGS7 was deleted in the RGS7Δ mutant (see text and Fig. 4) or swapped for the stretch of Gγ1 amino acids highlighted in Gγ1.