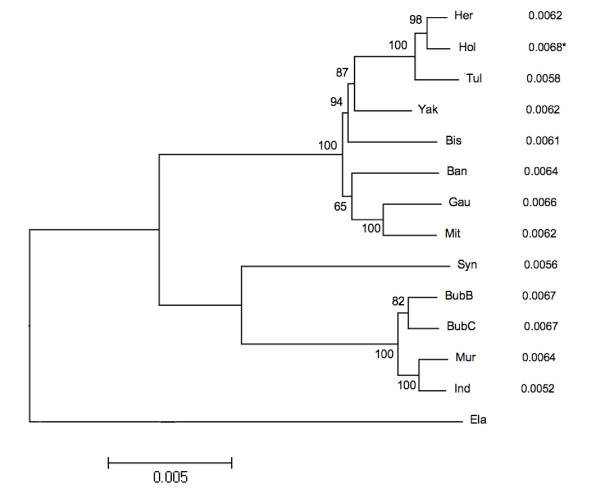
Figure 1.
Comparison of a neutral phylogeny and amino acid variation, neighbour joining analysis for members of the Bovini tribe (Anc: Ancient, Ban: Banteng, Bis: Bison, BubB & BubC: Asian buffalo, water and swamp type, Ela: Eland, Gau: Gaur, Her: Hereford, Ind & Mur: Indian water buffalo, Mit: Mithan, Hol: Holstein, Syn: African buffalo, Tul: Tuli, and Yak) using Kimura two parameter model with bootstrap values (5000 replicates) overlying branchpoints from an alignment of 21,055 bp with 1526 segregating sites from noncoding sequence, with the corresponding number of amino acid changes per site to the right of each species when compared to the Eland outgroup * highlights the largest number of aa changes per site.