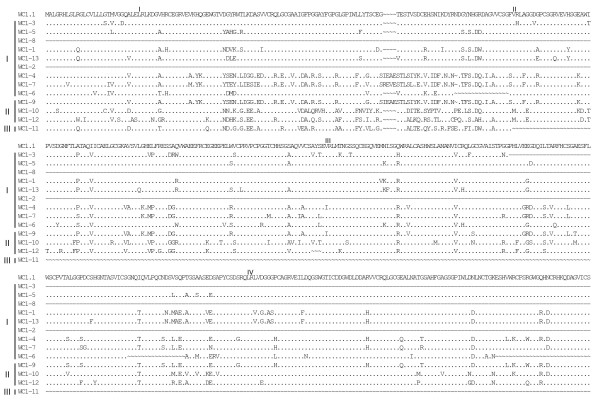
Figure 3.
Alignment of WC1 deduced amino acid sequences identified in the genome (part 1). Full-length deduced amino acid sequences of the annotated WC1 genes were aligned using ClustalW2 and the default parameters and were refined by hand. The archetypal WC1 sequence (WC1.1, GenBank accession number X63723) was included in the analysis for comparison. WC1-2, WC1-3, WC1-6, WC1-8 and WC1-12 sequences are partial due to gaps in the genome sequence. Gene types (I, II or III), as determined based on exon-intron structure are indicated to the left of the sequences. Identities are indicated by dots (.), gaps resulting from the alignment are indicated by tildes (~), gaps resulting from lack of genomic sequence (when the gaps were found adjacent and not within a coding region) are indicated by dashes (-). SRCR domains are indicated in roman numerals and the transmembrane region is shown underlined for the archetypal WC1.1 sequence.