Abstract
Undifferentiated nasopharyngeal carcinoma (NPC; WHO type III) is 100% associated with Epstein-Barr virus (EBV) infection and the fourth most prevalent cancer in Indonesian males. Therapy failure is high, since most patients come to the hospital at an advanced stage of disease. Screening for early-stage NPC is needed. Here, a simple and economical two-step enzyme-linked immunosorbent assay (ELISA) system is proposed for diagnosing NPC in high-risk populations, employing the peptide-based immunoglobulin A (IgA) EBNA1 plus viral capsid antigen p18 ELISA as an initial screening test and the IgA early antigen (EA) ELISA using a different set of EBV antigens as a confirmation test. A total of 151 NPC patients and 199 regional healthy EBV carriers were used to evaluate the two-step ELISA approach. Routinely, EBV IgG immunoblotting is used as a standard confirmation test. The sensitivity and specificity for diagnosing NPC by the two-step ELISA approach increased from 85.4% to 96.7% and 90.1% to 98%, respectively, with positive predictive values and negative predictive values increasing from 78.7 and 93.9% to 97.3 and 97.5%, respectively, relative to the immunoblotting confirmation system. On discrepant samples, additional testing was done by EBV DNA load quantification in blood. Results showed that 5/11 discrepant NPC samples with an elevated IgA EA ELISA also had elevated an EBV DNA load in the circulation (range, 3,200 to 25,820 copies/ml). Therefore, the IgA EA ELISA is proposed as a confirmation test in first-line NPC serological screening studies. This two-step EBV ELISA system provides a standardized approach for NPC screening and may be used in combination with dried blood sampling in future field studies for identification of early-stage NPC in high-risk regions.
Nasopharyngeal carcinoma (NPC) is a common cancer in China and Southeast Asia and closely associated with Epstein-Barr Virus (EBV) (26). In Indonesia, especially in the southern part of central Java, undifferentiated carcinoma (WHO type III) is the most common head and neck cancer and among the five most prevalent cancers overall. Due to unspecific symptoms and the hidden localization of the primary tumor at the early stage, more than 80% of the patients come to the hospital at a late stage (III or IV), when they already have metastasis in the cervical lymph node. Whereas late-stage disease has a poor prognosis and requires combined chemo-radiotherapy, early-stage NPC may reach complete remission by radiotherapy only (17). Therefore, screening for early-stage NPC among the population is important and clinically relevant. For developing countries, such an approach should be economical, employing standardization methods suited for mass screening.
Patients with NPC have high-level broad-spectrum anti-EBV antibodies, especially immunoglobulin A (IgA), compared to regional healthy carriers and patients with other head and neck diseases (13, 14). Our group recently demonstrated that the molecular diversity underlying anti-EBV IgG and IgA responses in NPC patients was different, requiring multiple EBV antigens for complete serological coverage (7). Prior studies in China and Taiwan have shown the feasibility of using IgA serology for population screening (2, 15, 27). However, in these studies laborious and poorly standardized cell-based serological techniques were used. Nevertheless, these studies revealed the appearance of serological abnormalities, i.e., positive EBV IgA responses 2 to 3 years prior to onset of NPC (2, 15), which clearly demonstrated the opportunity of using EBV serology for early-stage detection of NPC. This particularly applies for screening in high-risk groups, such as family members of NPC patients and patients with suspicious head and neck symptoms (18, 21).
For NPC serodiagnosis, cell-based indirect immunofluorescent assay (IFA) methods are still widely considered the gold standard. IFA involves the separate analysis of antibody responses to viral capsid antigen (VCA), early antigen (EA), and nuclear antigens (EBNA), each comprising multiple proteins and requiring different cell lines for specific analysis (10, 12, 13). However, this method shows considerable variation among laboratories and is time-consuming, subjective, and not suitable for large-scale automatic handling. Enzyme-linked immunosorbent assay (ELISA) techniques are increasingly used and have shown a better sensitivity and specificity compared to IFA and are suitable for large-scale application (4, 10, 11, 16, 20, 21).
Recently, we developed an EBV IgA ELISA based on a combination of VCA p18- and EBNA1-derived synthetic peptides which is routinely used as an NPC diagnostic test in our local hospital (Sardjito Hospital, Yogyakarta, Indonesia). This EBV IgA ELISA combines the separate features of IgA VCA and IgA EBNA1, each of which has its value in NPC diagnosis. The combination of these markers in a single assay provided sensitivity and specificity of 85.4% and 90.1%, respectively (8). The presence of NPC-related serological abnormalities can be confirmed by immunoblotting to reveal the spectrum of antibody responses, which has diagnostic value by itself (5, 7, 16, 25). The combined EBV IgA ELISA and immunoblot assay showed increased sensitivity and specificity and positive predictive value (PPV) and negative predictive value (NPV) of more than 95% (7, 8). Because immunoblot studies revealed a diagnostic value of multiple EBV proteins, in particular certain EBV-EA markers, we recently developed a separate IgA EA ELISA using native EA proteins (22). In addition to their role in primary diagnosis, anti-EBV IgA responses, in particular the IgA EA response, also have a distinct role for posttreatment follow-up monitoring, as declining responses correlate with a good prognosis and increasing responses are related to persistence of relapsing tumor (6, 16, 24). The availability of two distinct and biochemically well-defined EBV IgA ELISA systems addressing responses to different EBV antigens for NPC-specific serology may add to further standardization. In this study we evaluate the combination of these two tests for primary diagnosis of NPC in a high-incidence population in Indonesia.
MATERIALS AND METHODS
Sera.
Serum samples of NPC patients (n = 151) and healthy donors (n = 199) were obtained from the archives of the Gadjah Mada University, Yogyakarta, Indonesia. The NPC sera were taken on the first visit of patients. NPC staging was done by ear, nose, and throat examination and computer tomography (CT) scan and classified according to the 1997 Union International Cancer Control criteria. The NPC cases were confirmed by biopsy, with paraffin sections stained for EBV presence by standard Epstein Barr virus-encoded RNA-RNA in situ hybridization using a Peptide-based nucleic acid kit (Dako, Glostrup, Denmark) and by EBNA1 detection using monoclonal antibody OT1X (J. M. Middeldorp, VUMC, Amsterdam, The Netherlands). Sera from normal healthy donors were obtained from the local Red Cross blood bank.
Cell culture and cell extracts.
The P3HR1-derived cell line HH514.c16 (provided by G. Miller, Yale University, New Haven, CT) was grown at 37°C in flat-bottom or roller bottle culture, and cells were induced for expression of EA by culturing the cells at 1.5 × 106 cells/ml for 3 days at 32°C in the presence of 3 mM sodium butyrate, 20 ng/ml 12-O-tetradecanoylphorbol-1-acetate, and 0.5 mM phosphonoacetic acid. Purified nuclei from EA- or VCA-induced cells were prepared by Ficoll step gradient centrifugation exactly as described before (19). All cell culture chemicals were from Sigma, St. Louis, MO.
EBV antigens and EBV IgA ELISA.
The EA antigen used in this study was produced by 0.3 M salt extractions from purified EA nuclei as described elsewhere (22). Synthetic peptides for the EBNA1 plus VCA p18 ELISA ([EBNA1+VCA p18] ELISA) have been described before (7). Both ELISAs have been validated for NPC diagnosis in large panels of biopsy-confirmed NPC cases and regional controls, including blood bank donors and patients with non-NPC malignant and nonmalignant diseases presenting in the head and neck.
Standard ELISA plates (Greiner, Frickenhausen, Germany) were coated at 4°C overnight with 150 μl of a mixture of EBV VCA p18 and EBNA1 peptides or EA nuclear extract at 1 μg/ml in 50 mM sodium carbonate buffer, pH 9.6. Nonspecific binding was blocked for 1 h with phosphate-buffered saline (PBS)-3% bovine serum albumin at 37°C. Serum and conjugate incubations were for 1 h at 37°C followed by four washes with PBSt (PBS-0.1% [vol/vol] Triton-X, 1% bovine serum albumin). Human sera were diluted 1:100 in dilution buffer (PBSt, with an additional 1% normal rabbit serum for the IgA EA ELISA). Horseradish peroxidase-labeled rabbit anti-human IgA (Dako, Copenhagen, Denmark) was used at 1:4,000 in dilution buffer (with an additional 1% normal rabbit serum for the IgA EA ELISA), respectively. Horseradish peroxidase activity was detected by 3,3′,5,5′-tetramethylbenzidine (bioMerieux, Boxtel, The Netherlands) and stopped by adding 1 M H2SO4. The optical density at 450 nm (OD450) was determined and cutoff values (CoVs) were determined by receiver operating characteristic (ROC) analysis (8, 22).
Immunoblot assays.
In-house immunoblot assays using recombinant proteins or natural EBV antigen prepared from superinducible HH514 cells were done as described previously (7, 19, 22). Details and interpretation of immunoblot patterns have been described before (5, 7, 16, 25). For some experiments we used purified in-house recombinant proteins as described elsewhere (22).
Unfractionated whole-blood samples and EBV DNA load quantification by quantitative Light Cycler-based real-time PCR.
Unfractionated EDTA-anticoagulated whole-blood samples were obtained from NPC patients at the time of diagnosis and before any therapeutic intervention during their visit to Sardjito Hospital, Yogyakarta, Indonesia. Nucleic acid isolation and in-house Light Cycler-based real-time PCR procedures for EBV DNA load quantification, creating a 99-bp amplimer from a conserved region of the EBNA1-encoding BKRF1 gene, have been described elsewhere (23).
Data analysis.
All statistical analyses were done with the GraphPad Prism version 4.00 program (NJ). Cutoff values were determined by the ROC analysis based on the OD value of sera from 254 healthy donors and 151 NPC patients (3).
RESULTS
Distribution of study population.
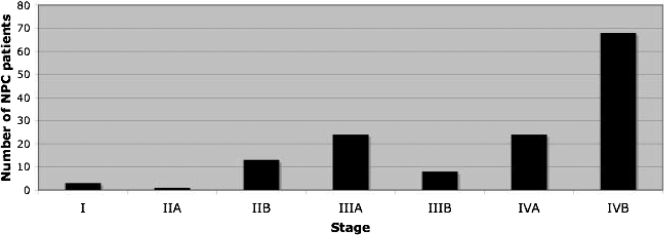
NPC patients were recruited prospectively during 2001 to 2003 at the Sardjito Hospital, Yogyakarta, Indonesia, at first visit prior to any therapeutic treatment. The male/female ratio was 2.3 (104/45). The age distribution ranged between 18 and 80 years with the majority between 31 and 60 years (mean, 46.8 years). Most patients (48%) were at stage IVB, and only 17/151 (11.3%) were at an early stage (I, IIA, or IIB) (Fig. 1).
FIG. 1.
Distribution of disease stage at presentation of 151 NPC patients, showing 11.3% were at an early stage (I, IIA, or IIB) and 88.7% were at a late stage (IIIA to IVB), with most of them (48%) being at stage IVB with distant metastasis.
Initial diagnosis using either IgA [EBNA1+VCA p18] or IgA EA ELISA.
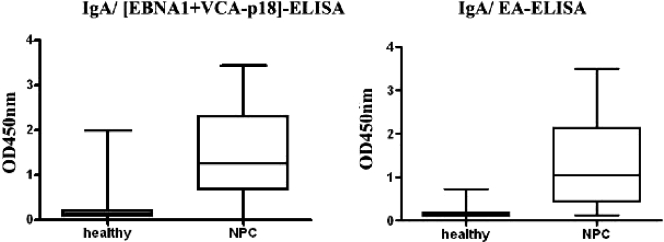
Analysis in healthy EBV carriers (n = 199) and NPC patients (n = 151) of serum IgA reactivity to [EBNA1+VCA p18] and EA using ELISA showed mean OD450 values of 0.19 and 0.16 for EBV carriers, whereas NPC patients yielded values of 1.48 and 1.39, respectively. The range of individual OD450 values for each ELISA are shown in Fig. 2. All healthy donors were EBV IgG seropositive, confirming their EBV carrier status (data not shown). The optimal CoV was defined by ROC analysis at 0.35 and 0.22 for [EBNA1+VCA p18] and EA, respectively (3). This yielded sensitivity and specificity values of 85.4% and 90.1% for the IgA [EBNA1+VCA p18] and 85.7% and 94% for the IgA EA ELISA.
FIG. 2.
Distribution frequency of EBV IgA ELISA values in the Indonesia panel for combination [EBNA1+VCA p18] antigen (A) and EA extract (A). Shown are the mean OD450 values and 25th and 75th percentiles of OD450 values for IgA reactivity in both ELISAs, using Indonesian serum panels consisting of healthy blood donors (n = 199) and NPC patients (n = 151). The cutoff values of both tests were 0.35 for IgA [EBNA1+VCA p18] and 0.22 for IgA EA. IgA reactivities to the [EBNA1+VCA p18] and EA ELISAs showed mean OD450 values of 0.19 and 0.16 for healthy EBV carriers (HC), and NPC patients had mean OD450 values of 1.48 and 1.39, respectively. These differences were highly significant (P < 0.0001).
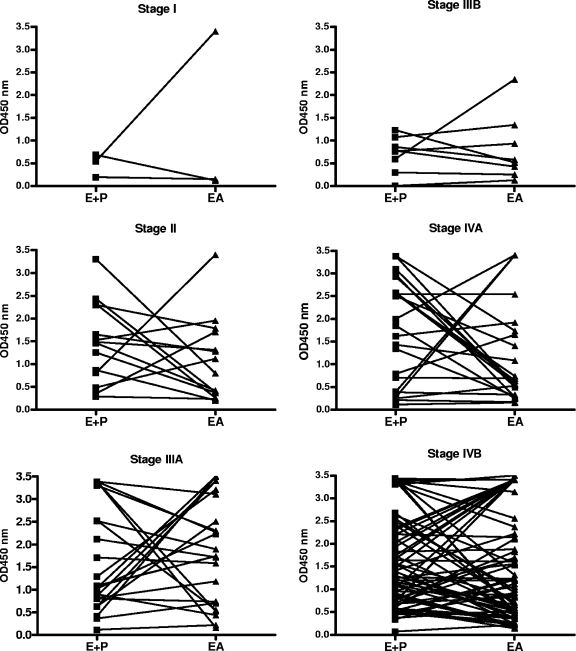
To illustrate that both ELISA systems detected different EBV IgA responses, the individual OD450 values in the IgA [VCA p18+EBNA1] ELISA and IgA EA ELISA for NPC cases at different stages were interconnected, revealing that although some sera showed similar reactivities in both ELISAs, generally a different level of response was detected (Fig. 3).
FIG. 3.
IgA [EBNA1+VCA p18] and IgA EA ELISA values for NPC patients at different stages of the disease. For each case, lines interconnect OD450 values in both ELISA systems. This reveals a generally different response level in either test, indicating that both ELISA systems detect distinct EBV IgA reactivities and may function for complementary serological testing.
Overall, 21/199 (10.6%) of healthy EBV IgG-seropositive blood donors were above the CoV in the peptide-based EBV IgA [EBNA1+VCA p18] ELISA, usually with rather low OD450 values (Fig. 2), and were initially classified as false positives. On the other hand, 15/151 (10%) confirmed NPC cases were at or below CoV in the EBV-IgA ELISA and were classified as false negatives in the initial screening test.
To further analyze false-positive and false-negative results, we performed EBV immunoblotting for IgG diversity. Nine of 21 false-positive healthy carriers had OD450 ELISA values in the gray area (i.e., within 0.20 of the CoV). All were tested for IgG diversity and showed a normal pattern, with six donors (pmi-3, pmi-67, pmi-88, pmi-108, pmi-154, and pmi-185) not revealing IgG EBV reactivity, in agreement with previous findings by Fachiroh et al. (7) (Table 1). Fifteen false-negative NPC sera were tested for IgG diversity by immunoblotting, and 10 of them gave an IgG high-diversity pattern (+3/+4), reflecting presence of NPC.
TABLE 1.
IgA EA ELISA values and IgG immunoblot scores for sera of healthy EBV carriers with low positive results for the IgA [EBNA1+VCA p18] ELISA
| Patient code | IgA ELISA (OD) fora:
|
IgG immunoblot diversity scoreb | |
|---|---|---|---|
| [EBNA1+VCA p18] (CoV, 0.354) | EA (CoV, 0.2162) | ||
| pmi-3 | 0.449* | 0.254** | − |
| pmi-9 | 0.541 | 0.171 | +1 |
| pmi-12 | 0.830 | 0.199 | +2 |
| pmi-17 | 0.490* | 0.158 | +1 |
| pmi-26 | 0.527 | 0.131 | +1 |
| pmi-29 | 0.441* | 0.109 | +1 |
| pmi-30 | 0.374* | 0.128 | +1 |
| pmi-43 | 0.463* | 0.150 | +1 |
| pmi-54 | 1.296 | 0.114 | +1 |
| pmi-67 | 0.364* | 0.135 | − |
| pmi-88 | 0.684 | 0.247** | − |
| pmi-91 | 1.462 | 0.143 | +1 |
| pmi-92 | 1.014 | 0.105 | +1 |
| pmi-95 | 0.532 | 0.210 | +1 |
| pmi-108 | 1.813 | 0.138 | − |
| pmi-152 | 0.492* | 0.044 | +1 |
| pmi-154 | 0.613 | 0.100 | − |
| pmi-173 | 0.395* | 0.067 | +1 |
| pmi-185 | 1.977 | 0.132 | − |
| pmi-194 | 0.418* | 0.266** | +1 |
| pmi-204 | 0.904 | 0.183 | +1 |
*, IgA [EBNA1+VCA p18] ELISA value in the gray area; **, IgA EA value above the cutoff.
IgG diversity score: −, negative; +1, normal; +2, normal reactive; +3 and +4, NPC pattern.
In our current routine NPC diagnosis at Sardjito hospital, because of the known level of false positivity, a positive IgA [EBNA1+VCA p18] reactivity is paralleled by immunoblot analysis for detection of abnormal IgG diversity, which is an effective but laborious method. Therefore, we here evaluated replacement of the immunoblotting with an IgA EA ELISA as a routine confirmation test.
IgA EA as the confirmation test of IgA [EBNA1+VCA p18].
Fifteen false-negative and 21 false-positive samples in the EBV IgA peptide-based screening ELISA (as defined above) were analyzed by immunoblotting to evaluate the IgG diversity to EBV proteins. The result revealed that 10/15 false negatives gave an IgG diversity pattern supportive of NPC, and all of the false-positive samples revealed a low or normal diversity pattern, supportive of uncomplicated EBV carriership. By IgA EA ELISA, 10/15 false negatives were identified as real NPC based on IgA EA values above the cutoff, and 8 of them were also confirmed by IgG immunoblotting. The IgA EA ELISA also revealed 19 of 22 false positives were confirmed as normal, based on IgA EA values below the cutoff (Tables 1 and 2). Therefore, the IgA EA ELISA can reliably replace immunoblotting as a confirmation assay. By using the IgA EA ELISA as the confirmation test, as shown in Table 3 the sensitivity and specificity of the two-step EBV IgA ELISA versus the one-step peptide-based ELISA for identification of true NPC cases increased from 85.4% to 96.7% and 90.1% to 98%, respectively, and PPV and NPV values increased from 78.7 and 93.9% to 97.3 and 97.5%, respectively.
TABLE 2.
EBV IgA ELISA value, IgG immunoblot result, and EBV DNA load of NPC cases with IgA-[EBNA1+VCAp18] below cutoff
| Sample code | IgA ELISA (OD) for:
|
Immunoblot diversity scorea | EBV DNA load/ml (CoV, 2,000) | |
|---|---|---|---|---|
| [EBNA1+ VCA p18] (CoV, 0.354) | EA (CoV, 0.216) | |||
| 01-2 | 0.253 | 0.530 | +3 | NAb |
| 01-3 | 0.274 | 3.407 | +4 | NA |
| 01-20 | 0.071 | 0.219 | +1 | NA |
| 01-46 | 0.213 | 0.167 | +1 | 200 |
| 01-56 | 0.201 | 0.155 | +3 | 0 |
| 02-11 | 0.338 | 1.118 | +4 | 23,400 |
| 02-37 | 0.081 | 1.182 | +4 | 7,600 |
| 02-48 | 0.115 | 0.217 | +3 | 3,200 |
| 02-79 | 0.288 | 0.236 | +2 | NA |
| 02-83 | 0.114 | 0.159 | +2 | 1,380 |
| 03-9 | 0.007 | 0.131 | +1 | 400 |
| 03-10 | 0.298 | 0.256 | +4 | 25,820 |
| 03-23 | 0.348 | 0.237 | +4 | 0 |
| 03-24 | 0.301 | 0.196 | +4 | 400 |
| 03-51 | 0.341 | 0.445 | +4 | 21,800 |
IgG diversity score: −, negative; +1, normal; +2, normal reactive; +3 and +4, NPC pattern.
NA, not available.
TABLE 3.
Diagnostic performance of EBV IgA ELISA for the Indonesia panel consisting of sera from 199 healthy EBV carriers and 151 NPC patients
| Test | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| IgA [EBNA1+VCA p18] | 85.4 | 90.1 | 78.7 | 93.9 |
| IgA EA | 85.7 | 94.0 | 79.4 | 94.9 |
| IgA [EBNA1+VCA p18] combined with IgA EA | 96.7 | 98 | 97.3 | 97.5 |
Confirmation of discrepant samples by EBV DNA load.
Six of 10 false negatives identified as possible NPC by IgA EA ELISA and from whom a whole-blood sample was available were tested for EBV DNA load in whole blood using the 99-bp EBNA1-based Light Cycler PCR as described before (23). Five of six had EBV copy numbers above the CoV (CoV, 2,000 copies/ml), ranging from 3,200 to 25,820 copies/ml (mean, 16,364 copies/ml), conforming their aberrant EBV activity and supportive of an NPC diagnosis.
DISCUSSION
In countries with a high NPC incidence, such as Indonesia, screening for early-stage disease is very important, since most patients currently come to the hospital at stage III or IV, with the consequences of therapy failure and a low survival rate posing considerable health care problems. Detection of EBV-related serological abnormalities, such as elevated EBV IgA levels, may provide a timely diagnosis of protracted early-stage NPC, as revealed in recent studies (2, 15). The availability of affordable yet accurate serological tests, which can be automated for large-scale applications, will be of benefit to cancer screening programs in developing countries. The IgA ELISA using defined and distinct EBV antigen may fulfill the criteria for such a screening approach, in particular when combined with simple sampling, such as dried blood collection, as shown by us recently (9). In that study, sera from well-defined groups of NPC patients and regional healthy individuals were used to evaluate the performance of a two-step ELISA system for detection and confirmation of NPC. The overall results showed that the two-step algorithm, using the peptide-based IgA [EBNA1+VCA p18] as the initial screening test and protein-based IgA EA as the confirmatory test, can provide highly sensitive and specific noninvasive detection of NPC.
The EBV IgA ELISA based on defined EBNA1 and VCA p18 synthetic peptide antigens combined in a single well fulfills the criteria for a well-standardized and cheap screening test. Peptides may be suitable replacements for natural or recombinant proteins as stable, reproducible, and cheap sources of antigen for an ELISA. Our data have shown that the peptide-based EBV IgA ELISA not only discriminates NPC patients from healthy EBV carriers (Fig. 2) but also from non-NPC tumor patients in a region with a high NPC prevalence (8). Importantly, the newly developed IgA EA ELISA addresses IgA responses to a distinct set of native EBV nuclear EA proteins as confirmed in this study (Fig. 3). Therefore, the combination of both IgA ELISAs provides additive independent serological information, contributing to improved diagnosis.
The combination of different technologies, like serology and EBV DNA load testing, was previously advised for diagnosis of NPC (20). EBV DNA load and serology are independent parameters, which means that they are not quantitatively related to each other. Therefore, the combination of these parameters will increase the diagnostic sensitivity (1, 23). Table 2 shows that NPC cases with low values for both EBV IgA ELISAs also have whole-blood EBV DNA loads below the CoV, and three of five with high EBV IgA responses also gave positive EBV DNA loads. This indicates that EBV DNA load may be used to confirm serology. However, EBV DNA load is positive in only some NPC patients and frequently is found at low levels. Furthermore, PCR techniques are cumbersome and require ultraclean lab facilities, which are difficult to realize and relatively expensive in developing countries.
In our hospital the EBV immunoblot assay, revealing the broad range (diversity) of IgG responses to EBV lytic proteins, is routinely used as a confirmation test, providing increased sensitivity and specificity and PPV and NPV values of greater than 95% for NPC diagnosis (8). However, immunoblot strip preparation and analysis are laborious. The immunoblot studies revealed the contribution of certain EA antigens as important markers of NPC-specific serology. Therefore, an independent serological test based on EA might be useful not only for confirmation but also for posttreatment prognostic monitoring in NPC (16, 22). Based on this study the IgA EA ELISA is proposed for the confirmation assay.
The two-step serological screening approach may permit identification of early-stage NPC cases by screening at-risk populations and thus contribute to improve early treatment and outcome of the disease. To confirm possible early-stage NPC in at-risk patients with positive EBV IgA results, a noninvasive nasopharyngeal brushing may be collected and examined for EBV DNA and RNA as described recently (24). Positive results directly reflect carcinogenic activity, in particular with detection of the carcinoma-specific EBV BARF1 mRNA. In a preliminary ongoing study at Sardjito Hospital, we are now screening random patients with chronic head and neck problems who are unresponsive to antibiotic treatment, and we have identified 2 true early-stage NPC cases among 30 patients analyzed using this two-step screening approach. If our work can confirm the early-stage diagnosis in larger populations, further expansion to family members of NPC patients and regional field hospitals is indicated.
Conclusions.
The two-step EBV IgA ELISA approach provides a reliable diagnostic format for NPC diagnosis and is proposed for screening of NPC in populations with high EBV prevalence, such as in Indonesia and other parts of Southeast Asia.
Acknowledgments
We thank the NPC team of Sardjito Hospital, Faculty of Medicine, Gadjah Mada University, Indonesia, for support in collecting patient samples and Bambang Hariwiyanto (ear, nose, and throat specialist) and A. Harijadi (pathologist) for providing clinical and pathological data. We also thank the EBV team in the Department of Pathology, Vrije Universiteit Medical Centre, Amsterdam, The Netherlands, for providing facilities and assistance.
This research was funded by The Netherlands Cancer Foundation (grant KWF-IN 2004-17) and by the European Union (grant Asia-link, contract no. ASI/B7-301/98/679-034).
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Chan, K. H., Y. L. Gu, F. Ng, P. S. Ng, W. H. Seto, J. S. Sham, D. Chua, W. Wei, Y. L. Chen, W. Luk, Y. S. Zong, and M. H. Ng. 2003. EBV specific antibody-based and DNA-based assays in serologic diagnosis of nasopharyngeal carcinoma. Int. J. Cancer 105706-709. [DOI] [PubMed] [Google Scholar]
- 2.Chien, Y. C., J. Y. Chen, M. Y. Liu, H. I. Yang, M. M. Hsu, J. Y. Chen, and C. S. Yang. 2001. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N. Engl. J. Med. 3451877-1882. [DOI] [PubMed] [Google Scholar]
- 3.Crowther, J. R. 2001. Validation of diagnostic test for infectious disease. Methods Mol. Biol. 149301-346. [Google Scholar]
- 4.Dardari, R., W. Hinderer, D. Lang, A. Benider, B. El Gueddari, I. Joab, A. Benslimane, and M. Khyatti. 2001. Antibody responses to recombinant Epstein-Barr virus antigens in nasopharyngeal carcinoma patients: complementary test of ZEBRA protein and early antigens p54 and p138. J. Clin. Microbiol. 393164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sanjose, S., R. Bosch, T. Schouten, S. A. Verkuijlen, A. Nieters, L. Foretova, M. Maynadie, P. L. Cocco, A. Staines, N. Becker, P. Brennan, Y. Benavente, P. Boffetta, C. J. Meijer, and J. M. Middeldorp. 2007. Epstein-Barr virus infection and risk of lymphoma: immunoblot analysis of antibody responses against EBV-related proteins in large series of lymphoma subjects and matched controls. Int. J. Cancer 1211806-1812. [DOI] [PubMed] [Google Scholar]
- 6.de Vathaire, F., H. Sancho-Garnier, H. de The, C. Pieddeloup, G. Schwab, J. H. Ho, R. Ellousz, C. Michaeu, M. Cammoun, Y. Cachin, and G. de The. 1988. Prognostic value of EBV markers in the clinical management of nasopharyngeal carcinoma (NPC): a multicenter follow-up study. Int. J. Cancer 42176-181. [DOI] [PubMed] [Google Scholar]
- 7.Fachiroh, J., T. Schouten, B. Hariwiyanto, D. K. Paramita, A. Harijadi, S. M. Haryana, M. H. Ng, and J. M. Middeldorp. 2004. Molecular diversity of Epstein-Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J. Infect. Dis. 19053-62. [DOI] [PubMed] [Google Scholar]
- 8.Fachiroh, J., D. K. Paramita, B. Hariwiyanto, A. Harijadi, H. L. Dahlia, S. R. Indrasari, H. Kusumo, Y. S. Zeng, T. Schouten, S. Mubarika, and J. M. Middeldorp. 2006. Single assay combination of Epstein-Barr virus (EBV) EBNA1 and viral capsid antigen p18-derived synthetic peptides for measuring anti-EBV immunoglobulin G (IgG) and IgA antibody levels in sera from nasopharyngeal carcinoma patients: option for field screening. J. Clin. Microbiol. 441459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fachiroh, J., P. R. Prasetyanti, D. K. Paramita, A. T. Prasetyawati, D. W. Anggrahini, S. M. Haryana, and J. M. Middeldorp. 2008. Dried-blood sampling for Epstein-Barr virus immunoglobulin G (IgG) and IgA serology in nasopharyngeal carcinoma screening. J. Clin. Microbiol. 461374-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gartner, B. C., J. M. Fischinger, K. Roemer, M. Mak, B. Fleurent, and N. Mueller-Lantzsch. 2001. Evaluation of recombinant line blot for diagnosis of Epstein-Barr virus compared with ELISA, using immunofluorescence as reference method. J. Virol. Methods 9389-96. [DOI] [PubMed] [Google Scholar]
- 11.Gartner, B. C., R. D. Hess, D. Bandt, A. Kruse, A. Rethwilm, K. Roemer, and N. Mueller-Lantzsch. 2003. Evaluation of four commercially available Epstein-Barr virus enzyme immunoassays with an immunofluorescence assay as the reference method. Clin. Diagn. Lab. Immunol. 1078-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henle, W., G. Henle, H. C. Ho, P. Burtin, Y. Cachin, P. Clifford, A. De Schryver, G. de The, V. Diehl, and G. Klein. 1970. Antibodies to Epstein Barr virus in nasopharyngeal carcinoma, other head and neck neoplasm group, and control group. J. Int. Cancer Inst. 44225-231. [PubMed] [Google Scholar]
- 13.Henle, G., and W. Henle. 1976. Epstein-Barr Virus specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int. J. Cancer 171-7. [DOI] [PubMed] [Google Scholar]
- 14.Ho, H. C., M. H. Ng, H. C. Kwan, and J. C. Chau. 1976. Epstein-Barr virus specific IgA and IgG serum antibodies in nasopharyngeal carcinoma. Br. J. Cancer 3455-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji, M. F., D. K. Wang, Y. L. Yu, Y. Q. Guo, J. S. Liang, W. M. Cheng, Y. S. Zong, K. H. Chan, S. P. Ng, W. I. Wei, D. T. T. Chua, J. S. T. Sham, and M. H. Ng. 2007. Sustained elevation of Epstein-Barr virus antibody levels preceding clinical onset of nasopharyngeal carcinoma. Br. J. Cancer 96623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karray, H., W. Ayadi, L. Fki, A. Hammami, J. Daoud, M. M. Drira, M. Frikha, R. Jlidi, and J. M. Middeldorp. 2005. Comparison of three different serological techniques for primary diagnosis and monitoring of nasopharyngeal carcinoma in two age groups from Tunisia. J. Med. Virol. 75593-602. [DOI] [PubMed] [Google Scholar]
- 17.Lin, J. C., J. S. Jan, C. Y. Hsu, W. M. Liang, R. S. Jiang, and W. Y. Wang. 2003. Phase III study of concurrent chemoradiotherapy versus radiotherapy alone for advanced nasopharyngeal carcinoma: positive effect on overall and progression-free survival. J. Clin. Oncol. 21631-637. [DOI] [PubMed] [Google Scholar]
- 18.Loh, K. S., B. C. Goh, J. Lu, W. S. Hsieh, and L. Tan. 2006. Familial nasopharyngeal carcinoma in a cohort of 200 patients. Arch. Otolaryngol. Head Neck Surg. 13282-85. [DOI] [PubMed] [Google Scholar]
- 19.Middeldorp, J. M., and P. Herbrink. 1988. Epstein-Barr virus specific marker molecules for early diagnosis of infectious mononucleosis. J. Virol. Methods 21133-146. [DOI] [PubMed] [Google Scholar]
- 20.Ng, W. T., T. K. Yau, R. W. Yung, W. M. Sze, A. H. Tsang, A. L. Law, and A. W. Lee. 2005. Screening for family members of patients with nasopharyngeal carcinoma. Int. J. Cancer 113998-1001. [DOI] [PubMed] [Google Scholar]
- 21.Ng, M. H., K. H. Chan, S. P. Ng, and Y. S. Zong. 2006. Epstein-Barr virus serology in early detection and screening of nasopharyngeal carcinoma. Chinese J. Cancer 25250-256. [PubMed] [Google Scholar]
- 22.Paramita, D. K., J. Fachiroh, W. T. Artama, E. van Benthem, S. M. Haryana, and J. M. Middeldorp. 2007. Native early antigen of Epstein-Barr virus, a promising antigen for diagnosis of nasopharyngeal carcinoma. J. Med. Virol. 791710-1721. [DOI] [PubMed] [Google Scholar]
- 23.Stevens, S. J. C., S. A. W. M. Verkuijlen, B. Hariwiyanto, Harijadi, J. Fachiroh, D. K. Paramita, I. B. Tan, S. M. Haryana, and J. M. Middeldorp. 2005. Diagnostic value of measuring Epstein-Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV Immunoglobulin A (IgA) and IgG antibody level in blood of nasopharyngeal carcinoma. J. Clin. Microbiol. 433066-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, S. J. C., S. A. W. M. Verkuijlen, M. C. Zwaan, and J. M. Middeldorp. 2006. Epstein-Barr virus (EBV) serology, but not EBV DNA load, for predicting distant metastases in a juvenile Caucasian nasopharyngeal carcinoma (NPC) patient without clinical response upon EBV lytic induction therapy. Head Neck 281040-1045. [DOI] [PubMed] [Google Scholar]
- 25.van Grunsven, W. M. J., A. Nabbe, and J. M. Middeldorp. 1993. Identification and molecular characterization of two diagnostically relevant marker proteins of the Epstein-Barr virus capsid antigen complex. J. Med. Virol. 40161-169. [DOI] [PubMed] [Google Scholar]
- 26.WHO International Agency for Cancer Research. 1997. Epstein-Barr virus, p. 47-373. IARC Monographs on the Evaluation of Carcinogenic Risks in Humans, publ. 70. IARC Press, Lyon, France. [PMC free article] [PubMed]
- 27.Zeng, Y., C. H. Gong, M. G. Jan, Z. Fun, L. G. Zhang, and H. Y. Li. 1983. Detection of Epstein-Barr virus IgA/EA antibody for diagnosis of nasopharyngeal carcinoma by immunoautoradiography. Int. J. Cancer 31599-601. [DOI] [PubMed] [Google Scholar]