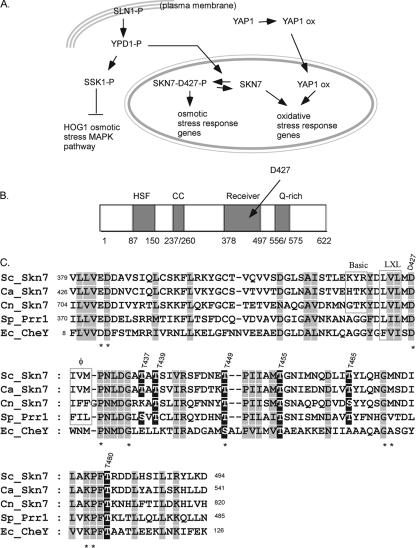
FIG. 1.
Skn7 function, structure, and sequence. (A) The role of the Skn7 transcription factor in two different signal transduction pathways. SLN1-P, YDP1-P, SSK1-P, and SKN7-D427-P are the phosphorylated forms of each protein. Skn7 is phosphorylated on the D427 residue in response to the activation of the SLN1 pathway (left), and Skn7 works with a second transcription factor, Yap1, to activate OXR genes (right). The OXR function of Skn7 is independent of SLN1 and the D427 residue. “ox” refers to unknown modifications to the Yap1 transcription factor in response to oxidative stress. (B) Domain structure of Skn7. HSF, the HSF-like N-terminal DBD of Skn7; CC, coiled-coil domain involved in some protein interactions; Receiver, domain that undergoes aspartyl phosphorylation in the response regulators of two-component-type phosphorelay pathways; Q-rich, C-terminal domain rich in glutamine. The amino acid residues comprising each domain are given below the cartoon. (C) ClustalW-based (38) sequence alignment between the receiver domain of four SKN7 homologs and the prokaryotic response regulator CheY. Ec_, Escherichia coli; Sc_, Saccharomyces cerevisiae; Sp_, Schizosaccharomyces pombe; Ca_, Candida albicans; and Cn_, Cryptococcus neoformans. The alignment was adjusted manually using the alignment editor GeneDoc (26). Columns consisting of identical or highly conserved amino acids are shown in gray. Residues with known importance in the response regulator family are marked with an asterisk. Conserved Ser/Thr residues are shown in white type on a black background. Dashed lines represent gaps in the alignment. Residues comprising a putative tripartite MAPK docking motif are boxed and labeled “Basic,” “LXL” (leucine, any amino acid residue, leucine), and “φ” (hydrophobic).