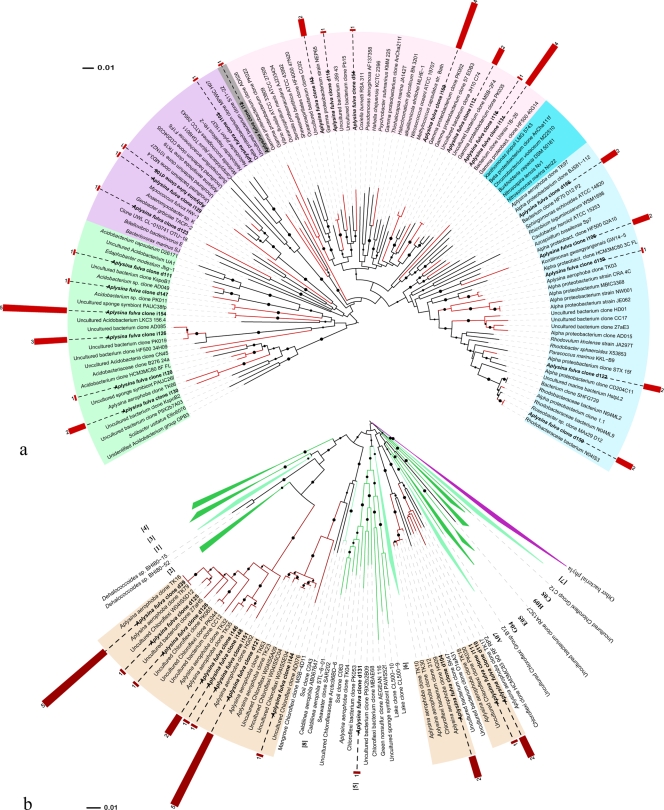
FIG. 5.
Phylogenetic analysis of bacterial 16S rRNA genes retrieved from A. fulva. The trees shown were inferred and drawn as explained in the legend to Fig. 3. Solid circles next to tree branches correspond to bootstrap values higher than 50%. A. fulva-derived clones are indicated in boldface, with their respective tree leaves marked in red. Trees display one representative clone per OTU. A single OTU embraces, here, all of the clone sequences sharing at least 99% similarity. Red bars next to A. fulva clone labels indicate the number of clones belonging to the corresponding OTU. (a) Evolutionary relationships of 125 taxa representative of the Acidobacteria (light green) and the alpha (light blue), beta (blue), gamma (light purple), and delta (purple) classes of Proteobacteria. There were 468 nucleotide positions in the final data set. (b) Phylogeny of the Chloroflexi as delineated by 167 representative taxa. All previously described Chloroflexi subdivisions—as designated by Rappé and Giovanonni (45a) (clades 1 to 8) and Costello and Schmidt (8a) (clades C12, C05, H09, E05, G04, B12, and A07)—are shown and appear, mostly, as collapsed clades. Clades composed exclusively by uncultured (dark green) or containing cultured (light green) representatives are differentiated. Four novel Chloroflexi subdivisions represented solely by sponge-derived uncultured bacteria are highlighted (colored ranges on sequence labels). There were a total of 528 aligned nucleotide positions in the final data set.